This is a preprint.
Human memory CD4+ T-cells recognize Mycobacterium tuberculosis-infected macrophages amid broader pathogen-specific responses
- PMID: 40060660
- PMCID: PMC11888249
- DOI: 10.1101/2025.02.23.639515
Human memory CD4+ T-cells recognize Mycobacterium tuberculosis-infected macrophages amid broader pathogen-specific responses
Update in
-
Human CD4+ T cells recognize Mycobacterium tuberculosis-infected macrophages amid broader responses.J Exp Med. 2025 Dec 1;222(12):e20250460. doi: 10.1084/jem.20250460. Epub 2025 Sep 24. J Exp Med. 2025. PMID: 40990918 Free PMC article.
Abstract
Recognition of macrophages infected with Mycobacterium tuberculosis (Mtb) is essential for CD4+ T cells to prevent tuberculosis (TB). Yet not all antigen-specific T cells recognize infected macrophages in human and murine models. Using monocyte-derived macrophages (MDMs) and autologous memory CD4+ T cells from individuals with latent Mtb infection (LTBI), we quantify T cell activation in response to infected macrophages. T cell antigen receptor (TCR) sequencing revealed >70% of unique and >90% of total Mtb-specific TCR clonotypes in stable LTBI are linked to recognition of infected macrophages, while a subset required exogenous antigen exposure, suggesting incomplete recognition. Clonotypes specific for multiple Mtb antigens and other pathogens were identified, indicating Mtb-specific and non-specific activation. Single-cell transcriptomics demonstrates Mtb-specific T cells express signature effector functions dominated by IFNγ, TNF, IL-2, and GM-CSF or chemokine production and signaling. We propose TB vaccines that elicit T cells capable of recognizing infected macrophages and expressing these canonical effector functions will offer protection against TB.
Keywords: GM-CSF; Mycobacterium tuberculosis; TB; TCR; bystander activation; human; infected macrophage; memory CD4 T cell; recognition.
Figures

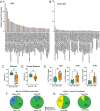

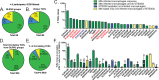



References
-
- Allan E.R.O., Tailor P., Balce D.R., Pirzadeh P., McKenna N.T., Renaux B., Warren A.L., Jirik F.R., and Yates R.M.. 2014. NADPH Oxidase Modifies Patterns of MHC Class II–Restricted Epitopic Repertoires through Redox Control of Antigen Processing. J. Immunol. 192:4989–5001. doi: 10.4049/jimmunol.1302896. - DOI - PubMed
-
- Antunes R. da S., Paul S., Sidney J., Weiskopf D., Dan J.M., Phillips E., Mallal S., Crotty S., Sette A., and Arlehamn C.S.L.. 2017. Definition of Human Epitopes Recognized in Tetanus Toxoid and Development of an Assay Strategy to Detect Ex Vivo Tetanus CD4+ T Cell Responses. PLoS ONE. 12:e0169086. doi: 10.1371/journal.pone.0169086. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
