This is a preprint.
Landscapes of missense variant impact for human superoxide dismutase 1
- PMID: 40060668
- PMCID: PMC11888409
- DOI: 10.1101/2025.02.25.640191
Landscapes of missense variant impact for human superoxide dismutase 1
Update in
-
Landscapes of missense variant impact for human superoxide dismutase 1.Am J Hum Genet. 2025 Oct 2;112(10):2295-2315. doi: 10.1016/j.ajhg.2025.08.019. Epub 2025 Sep 15. Am J Hum Genet. 2025. PMID: 40957416
Abstract
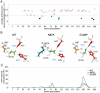
Amyotrophic lateral sclerosis (ALS) is a progressive motor neuron disease for which important subtypes are caused by variation in the Superoxide Dismutase 1 gene SOD1. Diagnosis based on SOD1 sequencing can not only be definitive but also indicate specific therapies available for SOD1-associated ALS (SOD1-ALS). Unfortunately, SOD1-ALS diagnosis is limited by the fact that a substantial fraction (currently 26%) of ClinVar SOD1 missense variants are classified as "variants of uncertain significance" (VUS). Although functional assays can provide strong evidence for clinical variant interpretation, SOD1 assay validation is challenging, given the current incomplete and controversial understanding of SOD1-ALS disease mechanism. Using saturation mutagenesis and multiplexed cell-based assays, we measured the functional impact of over two thousand SOD1 amino acid substitutions on both enzymatic function and protein abundance. The resulting 'missense variant effect maps' not only reflect prior biochemical knowledge of SOD1 but also provide sequence-structure-function insights. Importantly, our variant abundance assay can discriminate pathogenic missense variation and provides new evidence for 41% of missense variants that had been previously reported as VUS, offering the potential to identify additional patients who would benefit from therapy approved for SOD1-ALS.
Conflict of interest statement
Declaration of interests F.P.R. is an investor in Ranomics, Inc., and is an investor in and advisor for SeqWell, Inc., BioSymetrics, Inc., and Constantiam Biosciences, Inc.
Figures




References
-
- Johnston C.A., Stanton B.R., Turner M.R., Gray R., Blunt A.H.-M., Butt D., Ampong M.-A., Shaw C.E., Leigh P.N., and Al-Chalabi A. (2006). Amyotrophic lateral sclerosis in an urban setting: a population based study of inner city London: A population based study of inner city London. J. Neurol. 253, 1642–1643. - PubMed
-
- Pasinelli P., and Brown R.H. (2006). Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 7, 710–723. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
