This is a preprint.
The cerebrospinal fluid virome in people with HIV: links to neuroinflammation and cognition
- PMID: 40060671
- PMCID: PMC11888432
- DOI: 10.1101/2025.02.28.640732
The cerebrospinal fluid virome in people with HIV: links to neuroinflammation and cognition
Abstract
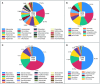
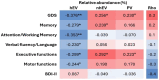
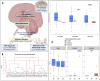
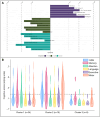
Despite effective HIV suppression, neuroinflammation and neurocognitive issues are prevalent in people with HIV (PWH) yet poorly understood. HIV infection alters the human virome, and virome perturbations have been linked to neurocognitive issues in people without HIV. Once thought to be sterile, the cerebrospinal fluid (CSF) hosts a recently discovered virome, presenting an unexplored avenue for understanding brain and mental health in PWH. This cross-sectional study analyzed 85 CSF samples (74 from PWH on suppressive antiretroviral therapy, and 11 from controls without HIV, CWH) through shotgun metagenomics for DNA/RNA viruses. Taxonomic composition (reads and contigs), α and β diversity, and relative abundance (RA) of prokaryotic (PV), human eukaryotic (hEV), and non-human eukaryotic viruses (nhEV) were evaluated in relation to HIV infection, markers of neuroinflammation and neurodegeneration, cognitive functions, and depressive symptoms. Sensitivity analyses and post-hoc cluster analysis on the RA of viral groups and blood-brain barrier permeability were also performed. Of 46 read-positive CSF samples, 93.5% contained PV sequences, 47.8% hEV, and 45.6% nhEV. Alpha diversity was lower in PWH versus CWH, although p>0.05. At β diversity analysis, HIV status explained 3.3% of the variation in viral composition (p=0.016). Contigs retained 13 samples positive for 8 hEV, 2 nhEV, and 6 PV. Higher RA of PV was correlated with higher CSF S100β (p=0.002) and β-Amyloid 1-42 fragment (βA-42, p=0.026), while higher RA of nhEV with poorer cognitive performance (p=0.022). Conversely, higher RA of hEV correlated with better cognition (p=0.003) and lower βA-42 (p=0.012). Sensitivity analyses in virome-positive samples only confirmed these findings. Three CSF clusters were identified and showed differences in astrocytosis, βA-42, tau protein, and cognitive functions. Participants with hEV-enriched CSF showed better cognitive performance compared to those with virus-devoid and nhEV-enriched CSF (models'p<0.05). This study provides the first comprehensive description of the CSF virome in PWH, revealing associations with neuroinflammation and cognition. These findings highlight the potential involvement of the CSF virome in brain health and inform about its composition, origin, and potential clinical implications in people with and without HIV.
Keywords: Bacteriophages; Brain microbiome; Central nervous system; Cognitive functions; Depression; Eukaryotic viruses; HIV-associated neurocognitive disorders; Prokaryotic viruses.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
