XGBMUT: Predicting the Functional Impact of Missense Mutations Using an Extreme Gradient Boost Classifier
- PMID: 40060867
- PMCID: PMC11886911
- DOI: 10.1021/acsomega.4c10179
XGBMUT: Predicting the Functional Impact of Missense Mutations Using an Extreme Gradient Boost Classifier
Abstract
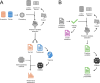
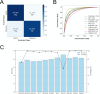
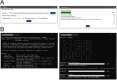
Millions of new mutations have been discovered largely due to advancements in genome projects, but characterizing their effects through traditional wet-lab experiments remains labor-intensive and time-consuming. Functional prediction algorithms offer a solution by enabling the efficient screening of mutations, thereby saving time and resources. The objective of this study was to develop a competitive algorithm for predicting the functional impact of missense mutations. A unified database and substitution matrices containing predictor variables specifically for missense mutations were initially constructed. Subsequently, values for the predictor variables were collected from the training and test sets derived from the ClinVar and HumsaVar databases. A series of supervised machine learning classifiers were then trained, and their performance was evaluated using the test set. The best-performing model was additionally compared against ten currently available functional prediction algorithms. The proposed algorithm, XGBMut, demonstrates exceptional accuracy in classifying missense mutations while also exhibiting a competitive performance. Additionally, a user-friendly graphical interface was developed to enhance accessibility for professionals in various fields. Unlike most existing methods, XGBMut eliminates the need for a web server dependency and the installation of third-party software, making it a more versatile tool for users.
© 2025 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Spencer D. H., Zhang B, Pfeifer J.. Single Nucleotide Variant Detection Using Next Generation Sequencing. Elsevier Inc.; 2014. doi:10.1016/B978-0-12-404748-8.00008-3. - DOI
-
- Kumar S U.; Kumar D T.; R S.; Doss C G. P.; Zayed H. An extensive computational approach to analyze and characterize the functional mutations in the galactose-1-phosphate uridyl transferase (GALT) protein responsible for classical galactosemia. Comput. Biol. Med. 2020, 117, 10358310.1016/j.compbiomed.2019.103583. - DOI - PubMed
-
- Richards S.; Aziz N.; Bale S.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine. 2015, 17 (5), 405–424. 10.1038/gim.2015.30. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
