Application of versatile reverse genetics system for feline coronavirus
- PMID: 40062768
- PMCID: PMC11960445
- DOI: 10.1128/spectrum.02692-24
Application of versatile reverse genetics system for feline coronavirus
Abstract
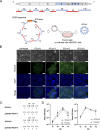
Feline infectious peritonitis (FIP) is a fatal disease caused by feline coronavirus (FCoV). Although multiple gene mutations in FCoV likely account for FIP pathogenesis, molecular studies for FCoV have been limited due to the lack of a suitable reverse genetics system. In the present study, we established a rapid PCR-based system to generate recombinant FCoV using the circular polymerase extension reaction (CPER) method for both serotype 1 and 2 viruses. Recombinant FCoV was successfully rescued at sufficient titers to propagate the progeny viruses with high sequence accuracy. The growth kinetics of recombinant FCoV were comparable to those of the parental viruses. We successfully generated recombinants harboring the spike gene from a different FCoV strain or a reporter HiBiT tag using the CPER method. The chimeric virus demonstrated similar characteristics with the parental virus of the spike gene. The reporter tag stably expressed after five serial passages in the susceptible cells, and the reporter virus could be applied to evaluate the sensitivity of antiviral inhibitors using the luciferase assay system to detect HiBiT tag. Taken together, our versatile reverse genetics system for FCoV shown herein is a robust tool to characterize viral genes even without virus isolation and to investigate the molecular mechanisms of the proliferation and pathogenicity of FCoV.
Importance: Feline infectious peritonitis is a highly fatal disease in cats caused by feline coronavirus variants that can infect systemically. Due to the lack of a versatile toolbox for manipulating the feline coronavirus genome, an efficient method is urgently needed to study the virus proteins responsible for the severe disease. Herein, we established a rapid reverse genetics system for the virus and demonstrated the capability of the recombinant viruses to be introduced with desired modifications or reporter genes without any negative impacts on virus characteristics in cell culture. Recombinant viruses are also useful to evaluate antiviral efficacy. Overall, our system can be a promising tool to reveal the molecular mechanisms of the viral life cycle of feline coronavirus and disease progression of feline infectious peritonitis.
Keywords: feline coronavirus; feline infectious peritonitis; reverse genetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Establishment of a Virulent Full-Length cDNA Clone for Type I Feline Coronavirus Strain C3663.J Virol. 2019 Oct 15;93(21):e01208-19. doi: 10.1128/JVI.01208-19. Print 2019 Nov 1. J Virol. 2019. PMID: 31375588 Free PMC article.
-
Establishment of Full-Length cDNA Clones and an Efficient Oral Infection Model for Feline Coronavirus in Cats.J Virol. 2021 Oct 13;95(21):e0074521. doi: 10.1128/JVI.00745-21. Epub 2021 Aug 18. J Virol. 2021. PMID: 34406859 Free PMC article.
-
Development and characterization of reverse genetics systems of feline infectious peritonitis virus for antiviral research.Vet Res. 2024 Sep 27;55(1):124. doi: 10.1186/s13567-024-01373-z. Vet Res. 2024. PMID: 39334482 Free PMC article.
-
Feline Coronaviruses: Pathogenesis of Feline Infectious Peritonitis.Adv Virus Res. 2016;96:193-218. doi: 10.1016/bs.aivir.2016.08.002. Epub 2016 Aug 31. Adv Virus Res. 2016. PMID: 27712624 Free PMC article. Review.
-
A Tale of Two Viruses: The Distinct Spike Glycoproteins of Feline Coronaviruses.Viruses. 2020 Jan 10;12(1):83. doi: 10.3390/v12010083. Viruses. 2020. PMID: 31936749 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

