Unveiling novel susceptibility genes and drug targets for basal cell carcinoma by a cross-tissue transcriptome-wide association study
- PMID: 40063313
- PMCID: PMC11893943
- DOI: 10.1007/s12672-025-02019-y
Unveiling novel susceptibility genes and drug targets for basal cell carcinoma by a cross-tissue transcriptome-wide association study
Abstract
Objective: To identify novel susceptibility genes and drug targets for basal cell carcinoma (BCC).
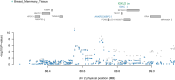
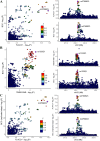
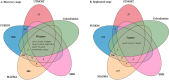
Methods: We performed a transcriptome-wide association study (TWAS) to identified the susceptibility genes and potential drug targets for BCC. The cross-tissue TWAS was conducted to discover the candidate genes for BCC. Functional Summary-based Imputation (FUSION) analysis was used to validate these genes in the single tissues. Multimarker Analysis of Genomic Annotation (MAGMA) was employed to further screen candidate genes. Summary data-based Mendelian randomization (SMR) and colocalization analyses were applied to infer causal relationships between candidate genes and BCC. The expression pattern of the identified genes in single-cell types was also investigated. Function, pathway enrichment and disease connection analyses were performed to understand the biological implication of identified genes. Additionally, druggability of the identified genes was evaluated to discover potential candidate drugs for BCC.
Results: Ninety-five genes were identified by cross-tissue TWAS analysis. Among them, 24 genes were confirmed by FUSION and MAGMA methods. Ten genes were further confirmed by SMR and colocalization analyses. Three genes were replicated by using another GWAS data. The potential interacting gene networks constructed with these identified genes were mainly involved in viral life cycle-HIV-1, GABAergic synapse, nicotine addiction, ether lipid metabolism, and mineral absorption pathways. AN-9 and amooranin might be candidate drugs for BCC.
Conclusions: We have identified 10 susceptibility genes associated with BCC risk, which might deepen our comprehension of BCC pathogenesis and illuminating new avenues for therapeutic and preventive drug development.
Keywords: Basal cell carcinoma; Cross-tissue TWAS; Drug targets; Susceptibility genes.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Ethical approval was waived because this study used the data from publicly available databases. Consent for publications: Not applicable. Competing interests: The authors declare no competing interests.
Figures






Similar articles
-
A cross-tissue transcriptome-wide association study reveals GRK4 as a novel susceptibility gene for COPD.Sci Rep. 2024 Nov 18;14(1):28438. doi: 10.1038/s41598-024-80122-w. Sci Rep. 2024. PMID: 39558015 Free PMC article.
-
Cross-Tissue Regulatory Network Analyses Reveal Novel Susceptibility Genes and Potential Mechanisms for Endometriosis.Biology (Basel). 2024 Oct 26;13(11):871. doi: 10.3390/biology13110871. Biology (Basel). 2024. PMID: 39596826 Free PMC article.
-
A cross-tissue transcriptome-wide association study identifies WDPCP as a potential susceptibility gene for coronary atherosclerosis.Atheroscler Plus. 2024 Nov 23;58:59-74. doi: 10.1016/j.athplu.2024.11.002. eCollection 2024 Dec. Atheroscler Plus. 2024. PMID: 39669798 Free PMC article.
-
A cross-tissue transcriptome-wide association study reveals novel susceptibility genes for migraine.J Headache Pain. 2024 Jun 5;25(1):94. doi: 10.1186/s10194-024-01802-6. J Headache Pain. 2024. PMID: 38840241 Free PMC article.
-
From GWAS to Gene: Transcriptome-Wide Association Studies and Other Methods to Functionally Understand GWAS Discoveries.Front Genet. 2021 Sep 30;12:713230. doi: 10.3389/fgene.2021.713230. eCollection 2021. Front Genet. 2021. PMID: 34659337 Free PMC article. Review.
References
-
- Heath M, Bar A. Basal cell carcinoma. Dermatol Clin. 2023;41(1):13–21. - PubMed
-
- Kim D, Kus K, Ruiz E. Basal cell carcinoma review. Hematol Oncol Clin North Am. 2019;33(1):13–24. - PubMed
-
- Peris K, Fargnoli M, Kaufmann R, Arenberger P, Bastholt L, Seguin N, et al. European consensus-based interdisciplinary guideline for diagnosis and treatment of basal cell carcinoma—update 2023. Eur J Cancer. 2023;192:113254. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
