The Spectrum of IDH- and H3-Wildtype High-Grade Glioma Subgroups Occurring across Teenage and Young Adult Patient Populations
- PMID: 40063504
- PMCID: PMC12314528
- DOI: 10.1158/1078-0432.CCR-24-1256
The Spectrum of IDH- and H3-Wildtype High-Grade Glioma Subgroups Occurring across Teenage and Young Adult Patient Populations
Abstract
Purpose: High-grade gliomas (HGG) occur in any central nervous system location and at any age. HGGs in teenagers/young adults (TYA) are understudied. This project aimed to characterize these tumors to support accurate stratification of patients.
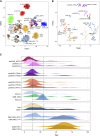
Experimental design: 207 histone/IDH wild-type tumors from patients aged 13 to 30 years were collected. DNA methylation profiling [Illumina EPIC BeadArrays, brain tumor classifier (MNPv12.8 R package)] classified cases against reference cohorts of HGG. Calibrated scores guided characterization workflows [RNA-based ArcherDx fusion panel (n = 92), whole-exome sequencing (n = 107), and histology review).
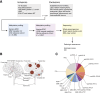
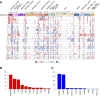
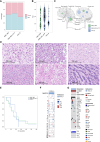
Results: 53.4% (n = 86) matched as pediatric-type subgroups [pedHGG_RTK1A/B/C (31.7%, n = 51, PDGFRA, CDKN2A/B, SETD2, and NF1 alterations), pedHGG_MYCN (8.1%, n = 13, MYCN/ID2 amplifications), and pedHGG_RTK2A/B (7.5%, n = 12, TP53, BCOR, ATRX, and EGFR alterations)]. Eighteen percent (n = 29) classified as adult-type subgroups [GBM_MES (15.5%, n = 25, enriched for RB1, PTEN, and NF1 alterations) and GBM_RTK1/2 (2.5%, n = 4, CDK4 amplifications)]. Twenty-three cases (14.7%) classified as novel, poorly characterized subgroups with distinct methylation profiles and molecular features [pedHGG_A/B (n = 10 6.2%), HGG_E (n = 6 3.7%), HGG_B (n = 2 1.0%), and GBM_CBM (n = 5 3.1%)] with variable histologic morphology. Eight cases (5.1%) showed hypermutator phenotypes, enriched in HGG_E, one of which was associated with constitutional mismatch repair deficiency, and their sibling, who was diagnosed with the same syndrome, was diagnosed with a tumor that classified as a pedHGG_RTK1B. HGGs that have developed on a background of previous treatment for a childhood cancer are detected in the TYA population, classifying most frequently as pedHGG_RTK1 and contributing to the poor prognosis of this subgroup. Age distribution/molecular profile comparisons using publicly available methylation/sequencing data (and from local diagnostic cohorts) for HGG_B (n = 19), GBM_CBM (n = 35), and GBM_MES_ATYP (n = 102), irrespective of age, show that HGG_B is a TYA-specific subgroup (median age 29 years) and that GBM_CBM and GBM_MES_ATYP show a peak of distribution in the TYA population but also have a wider age distribution (median age 35.7 and 50.5 years, respectively), with the latter showing distinct differences in copy-number profiles compared with older adults in the same subgroup and containing fewer chr7 gains, chr10 losses, more CDKN2A/B deletions and MET amplifications, and a worse survival compared with adult-specific GBM_MES_TYP.
Conclusions: TYA HGGs comprise novel methylation subgroups with distinct methylation and molecular profiles. Accurate stratification of these patients will open opportunities to more effective treatments, including immune checkpoint, MAPK pathway, and PDGFRA inhibitors. See related commentary by Ritzmann et al., p. 3110.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
J. Sidpra reports grants and personal fees from Cancer Research UK, grants from Olivia Hodson Foundation, and personal fees from University College London outside the submitted work. D.S. Ziegler reports grants from Accendatech and personal fees from Medison Pharma, Roche, Novartis, Alexion, FivepHusion, Amgen, AstraZeneca, Bayer, and Day One Therapeutics outside the submitted work. T.S. Jacques reports grants from the National Institute for Health and Care Research during the conduct of the study as well as grants from The Brain Tumor Charity, Cancer Research UK, Chan Zuckerberg Initiative, Children with Cancer UK, and Olivia Hodson Cancer Fund and other support from Repath Ltd, Neuropath Ltd, and Neuropathology and Applied Neurobiology outside the submitted work. D. Hargrave reports personal fees and nonfinancial support from Day One Therapeutics and Novartis; grants, personal fees, and nonfinancial support from AstraZeneca; and personal fees from Ipsen and Biodexa outside the submitted work. L. Marshall reports personal fees from Bayer, Eisai, and Merck outside the submitted work. No disclosures were reported by the other authors.
Figures


References
-
- Giangaspero F, Gianno F, Antonelli M, Ferretti E, Massimino M, Arcella A. Pediatric high-grade glioma: a heterogeneous group of neoplasms with different molecular drivers. Glioma 2018;1:117.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

