Unidirectional recruitment between MeCP2 and KSHV-encoded LANA revealed by CRISPR/Cas9 recruitment assay
- PMID: 40063648
- PMCID: PMC11913271
- DOI: 10.1371/journal.ppat.1012972
Unidirectional recruitment between MeCP2 and KSHV-encoded LANA revealed by CRISPR/Cas9 recruitment assay
Abstract
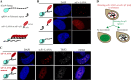
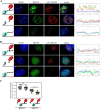
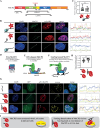
Kaposi's sarcoma-associated herpesvirus (KSHV, HHV-8) is associated with several human malignancies. During latency, the viral genomes reside in the nucleus of infected cells as large non-integrated plasmids, known as episomes. To ensure episome maintenance, the latency protein LANA tethers the viral episomes to the cell chromosomes during cell division. Directional recruitment of protein complexes is critical for the proper function of many nuclear processes. To test for recruitment directionality between LANA and cellular proteins, we directed LANA via catalytically inactive Cas9 (dCas9) to a repeat sequence to obtain easily detectable dots. Then, the recruitment of nuclear proteins to these dots can be evaluated. We termed this assay CRISPR-PITA for Protein Interaction and Telomere Recruitment Assay. Using this protein recruitment assay, we found that LANA recruits its known interactors ORC2 and SIN3A. Interestingly, LANA was unable to recruit MeCP2, but MeCP2 recruited LANA. Both LANA and histone deacetylase 1 (HDAC1) interact with the transcriptional-repression domain (TRD) and the methyl-CpG-binding domain (MBD) of MeCP2. Similar to LANA, HDAC1 was unable to recruit MeCP2. While heterochromatin protein 1 (HP1), which interacts with the N-terminal of MeCP2, can recruit MeCP2. We propose that available interacting domains force this recruitment directionality. We hypothesized that the tandem repeats in the SunTag may force MeCP2 dimerization and mimic the form of DNA-bound MeCP2. Indeed, providing only the tandem epitopes of SunTag allows LANA to recruit MeCP2 in infected cells. Therefore, CRISPR-PITA revealed the rules of unidirectional recruitment and allowed us to break this directionality.
Copyright: © 2025 Lavi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Rainbow L, Platt GM, Simpson GR, Sarid R, Gao SJ, Stoiber H, et al.. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71(8):5915–21. doi: 10.1128/JVI.71.8.5915-5921.1997 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

