DNA lesions can frequently precede DNA:RNA hybrid accumulation
- PMID: 40064914
- PMCID: PMC11893903
- DOI: 10.1038/s41467-025-57588-x
DNA lesions can frequently precede DNA:RNA hybrid accumulation
Abstract
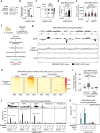
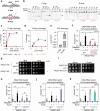
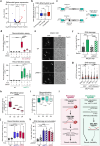
While DNA:RNA hybrids contribute to multiple genomic transactions, their unscheduled formation is a recognized source of DNA lesions. Here, through a suite of systematic screens, we rather observed that a wide range of yeast mutant situations primarily triggering DNA damage actually leads to hybrid accumulation. Focusing on Okazaki fragment processing, we establish that genic hybrids can actually form as a consequence of replication-born discontinuities such as unprocessed flaps or unligated Okazaki fragments. Strikingly, such "post-lesion" DNA:RNA hybrids neither detectably contribute to genetic instability, nor disturb gene expression, as opposed to "pre-lesion" hybrids formed upon defective mRNA biogenesis, e.g., in THO complex mutants. Post-lesion hybrids similarly arise in distinct genomic instability situations, triggered by pharmacological or genetic manipulation of DNA-dependent processes, both in yeast and human cells. Altogether, our data establish that the accumulation of transcription-born DNA:RNA hybrids can occur as a consequence of various types of natural or pathological DNA lesions, yet do not necessarily aggravate their genotoxicity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
- FDT202304016590/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
- EQU202003010245/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
- PJA20181208112/Fondation ARC pour la Recherche sur le Cancer (ARC Foundation for Cancer Research)
- ANR-18-CE12-003/Agence Nationale de la Recherche (French National Research Agency)
- RS21/75-21/Ligue Contre le Cancer
- ANR-11-LABX-0071/Agence Nationale de la Recherche (French National Research Agency)
- ANR-18-IDEX-0001/Agence Nationale de la Recherche (French National Research Agency)
- GM50237/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R35 GM118180/GM/NIGMS NIH HHS/United States
- ANR-21-CE12-0040/Agence Nationale de la Recherche (French National Research Agency)
LinkOut - more resources
Full Text Sources

