SNPeBoT: a tool for predicting transcription factor allele specific binding
- PMID: 40065237
- PMCID: PMC11895208
- DOI: 10.1186/s12859-025-06094-4
SNPeBoT: a tool for predicting transcription factor allele specific binding
Abstract
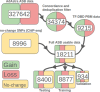
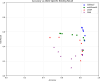
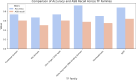
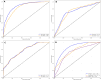
Background: Mutations in non-coding regulatory regions of DNA may lead to disease through the disruption of transcription factor binding. However, our understanding of binding patterns of transcription factors and the effects that changes to their binding sites have on their action remains limited. To address this issue we trained a Deep learning model to predict the effects of Single Nucleotide Polymorphisms (SNP) on transcription factor binding. Allele specific binding (ASB) data from Chromatin Immunoprecipitation sequencing (ChIP-seq) experiments were paired with high sequence-identity DNA binding Domains assessed in Protein Binding Microarray (PBM) experiments. For each transcription factor a paired DNA binding Domain was selected from which we derived E-score profiles for reference and alternate DNA sequences of ASB events. A Convolutional Neural Network (CNN) was trained to predict whether these profiles were indicative of ASB gain/loss or no change in binding. 18211 E-score profiles from 113 transcription factors were split into train, validation and test data. We compared the performance of the trained model with other available platforms for predicting the effect of SNP on transcription factor binding. Our model demonstrated increased accuracy and ASB recall in comparison to the best scoring benchmark tools.
Conclusion: In this paper we present our model SNPeBoT (Single Nucleotide Polymorphism effect on Binding of Transcription Factors) in its standalone and web server form. The increased recovery and prediction accuracy of allele specific binding events could prove useful in discovering non-coding mutations relevant to disease.
Keywords: Gene regulation; Neural network; Transcription factor.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Conflict Of Interest: The authors declare no competing interests.
Figures

References
-
- Cavalli M, Pan G, Nord H, Wallerman O, Wallén Arzt E, Berggren O, Elvers I, Eloranta ML, Rönnblom L, Lindblad Toh K, Wadelius C. Allele-specific transcription factor binding to common and rare variants associated with disease and gene expression. Hum Genet. 2016;135(5):485–97. 10.1007/s00439-016-1654-x. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

