Solvation, geometry, and assembly of the tobacco mosaic virus
- PMID: 40065976
- PMCID: PMC11891515
- DOI: 10.1093/pnasnexus/pgaf065
Solvation, geometry, and assembly of the tobacco mosaic virus
Abstract
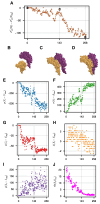
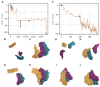
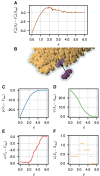
Biological self-assembly is a fundamental aspect in the development of complex structures in nature. A paradigm for such a process is the assembly of tobacco mosaic virus (TMV) capsid proteins into helical rods around the viral genome. The self-assembly process of the virus is typically modelled through attractive interactions between protein subunits, however capsid proteins also interact with their aqueous environment through solvation free energy. An open question is what role solvation plays in virus self-assembly. Here, we show that a purely geometric model of nonpolar solvation free energy, the morphometric approach, is sufficient to simulate the assembly of up to three protein subunits of TMV. The lowest solvation free energy states we find in a geometric simulation setting are remarkably close to the correctly assembled states of various experimentally determined structures. This demonstrates that van der Waals forces and entropic considerations are sufficient to guide assembly in the absence of attractive interactions between protein subunits. It further illustrates the impact of the morphometric approach as a computationally efficient model for nonpolar solvation free energy of solutes, in particular those with complicated geometry. This demonstration of the role of solvation raises important questions about the driving forces behind biological self-assembly and the paramount role of geometry.
Keywords: geometry; morphometric energy; self-assembly; solvation; tobacco mosaic virus.
© The Author(s) 2025. Published by Oxford University Press on behalf of National Academy of Sciences.
Figures



Similar articles
-
Coordinated two-disk nucleation, growth and properties, of virus-like particles assembled from tobacco-mosaic-virus capsid protein with poly(A) or oligo(A) of different length.Eur J Biochem. 1984 Apr 2;140(1):119-27. doi: 10.1111/j.1432-1033.1984.tb08074.x. Eur J Biochem. 1984. PMID: 6705790
-
Assembly of tobacco mosaic virus and TMV-like pseudovirus particles in Escherichia coli.Arch Virol Suppl. 1994;9:543-58. doi: 10.1007/978-3-7091-9326-6_52. Arch Virol Suppl. 1994. PMID: 7518274
-
Nanoparticle encapsidation of Flock house virus by auto assembly of Tobacco mosaic virus coat protein.Int J Mol Sci. 2014 Oct 14;15(10):18540-56. doi: 10.3390/ijms151018540. Int J Mol Sci. 2014. PMID: 25318056 Free PMC article.
-
Molecular assembly of tobacco mosaic virus in vitro.Adv Biophys. 1986;22:95-149. doi: 10.1016/0065-227x(86)90004-3. Adv Biophys. 1986. PMID: 3551520 Review.
-
Switching in the self-assembly of tobacco mosaic virus.Adv Biophys. 1990;26:157-85. doi: 10.1016/0065-227x(90)90011-h. Adv Biophys. 1990. PMID: 2082726 Review.
References
-
- Caspar DLD. Assembly and stability of the tobacco mosaic virus particle. In: Advances in Protein Chemistry. Vol. 18. Academic Press, 1964. p. 37–121. https://www.sciencedirect.com/science/article/pii/S0065323308602685. 10.1016/S0065-3233(08)60268-5 - DOI - PubMed
-
- Durham ACH, Klug A. 1971. Polymerization of tobacco mosaic virus protein and its control. Nat New Biol. 229(2):42–46. - PubMed
LinkOut - more resources
Full Text Sources

