Cell-free DNA for detection and monitoring of extramedullary AML relapse
- PMID: 40066001
- PMCID: PMC11891922
- DOI: 10.1002/hem3.70097
Cell-free DNA for detection and monitoring of extramedullary AML relapse
Abstract
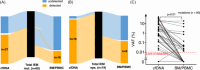
Isolated extramedullary manifestations (IEM) of acute myeloid leukemia (AML) are recurrent events, especially following allogeneic hematopoietic cell transplantation (alloHCT). To date, measurable residual disease (MRD) assessment for this difficult-to-treat patient cohort has not been established. In this study, we evaluated highly sensitive next-generation sequencing (NGS) of IEM-AML tumor and compared it with cell-free DNA (cfDNA) from plasma, as well as highly sensitive NGS analysis of bone marrow mononuclear cells (BMMC) and peripheral blood mononuclear cells (PBMC), in a cohort of 15 IEM-AML patients with 19 IEM-AML episodes. cfDNA demonstrated a superior representation of IEM-AML tumor mutations compared to BMMC or PBMC, with a median variant allele frequency (VAF) of 0.8% and a mutation detection rate of 62% (37 of 60 mutations), compared to a median VAF of 0.05% and detection rate of 27%, respectively (16 of 60 mutations, p < 0.01). Among 44 mutations identified in 14 IEM-AML relapse tumors, 30 mutations (68%) were known from initial diagnosis. Using diagnostic mutations from initial diagnosis for MRD analysis and detection of IEM-AML relapse, 16 of 17 IEM-AML relapse episodes were detected via cfDNA, whereas only 7 of 17 were identified using conventional analysis of BMMC or PBMC. Our findings demonstrate that cfDNA analysis from plasma effectively captures the molecular profile of IEM-AML. More than one-third of clinically relevant mutations were exclusively detected through cfDNA and were missed by conventional NGS-MRD of BMMC or PBMC. These results suggest that MRD monitoring using cfDNA offers a more comprehensive and sensitive approach to detecting IEM-AML relapse compared to standard methods.
© 2025 The Author(s). HemaSphere published by John Wiley & Sons Ltd on behalf of European Hematology Association.
Conflict of interest statement
M. H. declares honoraria from Astellas, Daiichi Sankyo, Janssen, Miltenyi, Otsuka, Qiagen, and Servier; paid consultancy for Abbvie, AvenCell, Ascentage Pharma, Bristol Myers Squibb, Janssen, Jazz Pharmaceuticals, LabDelbert, Novartis, Pfizer, and Servier; and research funding to his institution from Abbvie, Bayer Pharma AG, Jazz Pharmaceuticals, Glycostem, Karyopharm, PinotBio, Servier, and Toray. S. B. received a speaker honorary from ThermoFisher. U. L. declares honoraria and travel support from AstraZeneca, BMS, GSK, Menarini Stemline, Novartis, QuIP, and Servier. F. H. H. served as an advisor for Novartis, CTI, Celgene/BMS, Janssen, Abbvie, GSK, Merck, and AOP and received research funding from Novartis, Celgene/BMS, and CTI. N. D. D. declares research funding to her institution from Illumina. The remaining authors declare no conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
