Unveiling SSR4: a promising biomarker in esophageal squamous cell carcinoma
- PMID: 40066443
- PMCID: PMC11891195
- DOI: 10.3389/fimmu.2025.1544154
Unveiling SSR4: a promising biomarker in esophageal squamous cell carcinoma
Abstract
Background: Esophageal squamous cell carcinoma (ESCC) represents a frequent cancer with a poor prognosis. Altered glucose metabolism contributes factor to ESCC progression. In our previous study, signal sequence receptor subunit delta (SSR4) was included in an ESCC prognostic model; however, the mechanisms underlying SSR4 implication in ESCC remain ambiguous. Accordingly, we aim to determine the interconnection between SSR4 expression and clinical characteristics of ESCC.
Methods: This differential expression and prognostic significance of SSR4 was performed using bulk RNA-seq data and 110 patients with complete follow-up information. The ESCC cell subsets with the highest gene expression levels were identified with single-cell data. Gene function and enrichment, immune infiltration, cell communication, and molecular docking analyses were performed.
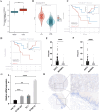
Results: Unlike adjacent non-cancerous tissues, SSR4 was overexpressed in ESCC tissues, validated by both reverse transcription-qPCR and IHC staining. SSR4 expression was related to the N stage, lymph node metastasis, and AJCC TNM classification stage. Patients exhibiting low SSR4 expression had a more favorable prognosis. The highest SSR4 expression was recognized in tumor plasma cells. Continued exploration of immune infiltration highlighted a close association between SSR4 gene expression and the infiltration of immune cells such as plasma cells. On dividing cells into SSR4-positive and -negative groups, CellChat analysis indicated that SSR4 may regulate the interactions that existed between ESCC tumor plasma cells and the tumor microenvironment (TME) by modulating the MIF/CD74/CXCR4 axis.
Conclusion: The SSR4 gene may have significant relevance with clinical pathological factors, and play a critical role in the regulation of tumor microenvironment of ESCC patients. Overall, SSR4 may be a promising ESCC biomarker with prospective applicability in clinical diagnosis as well as the development of targeted treatment approaches in patients of ESCC.
Keywords: SSR4; biomarker; esophageal squamous cell carcinoma; immune cells; single-cell transcriptomics.
Copyright © 2025 Zhang, Jia, Li, Song and Gong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

