The influence of genetic polymorphisms on cytokine profiles in pediatric COVID-19: a pilot study
- PMID: 40066463
- PMCID: PMC11891370
- DOI: 10.3389/fped.2025.1523627
The influence of genetic polymorphisms on cytokine profiles in pediatric COVID-19: a pilot study
Abstract
Introduction: Recent studies have underscored the importance of genetic factors in predicting COVID-19 susceptibility and severity. While cytokine storms are crucial in disease severity, genetic predisposition significantly influences immune responses. Our study examined genes related to SARS-CoV-2 invasion (ACE2 rs2074192) and interferon-induced immunity (IFNAR2 rs2236757, TYK2 rs2304256, OAS1 rs10774671, OAS3 rs10735079). Additionally, we investigated genes linked to Kawasaki disease (CD40 rs4813003, FCGR2A rs1801274, CASP3 rs113420705) that play roles in immunogenesis.
Methods: The pilot study, which involved 75 pediatric patients aged one month to 17 years [43 patients with active COVID-19, 17 children with multisystem inflammatory syndrome in children (MIS-C), and 15 healthy controls], was conducted in Ternopil, Ukraine. Gene polymorphism was studied for all patients. ELISA kits were used for interleukin studies, including Human IL-1β (Interleukin 1 Beta), Human IL-6 (Interleukin 6), Human IL-8 (Interleukin 8), Human IL-12 (Interleukin 12), Human IFN-α (Interferon Alpha), and Human TNF-α (Tumor Necrosis Factor Alpha). Statistical analysis was performed using IBM SPSS Statistics 21 and GraphPad Prism 8.4.3.
Results: The analysis identified significant gene-cytokine associations in pediatric COVID-19 patients. The ACE2 rs2074192 T allele correlated with increased IL-1β, IL-6, IL-8, and TNF-α. The IFNAR2 rs2236757 A allele was linked to elevated IL-1β and IL-12 levels and low IFN-α levels, while OAS1 rs10774671 A allele carriers also exhibited lower IFN-α levels. OAS1 rs10774671 was prognostically crucial for determining IL-8 levels in children infected with SARS-CoV-2. OAS3 gene polymorphism rs10735079 was associated with changes in IL-6 levels, precisely a high level. The CD40 rs4813003 T allele increased IFN-α levels, while carriers of allele C had higher levels of IL-12. The results of our study revealed a correlation between IL-8 levels and the FCGR2A gene polymorphism rs1801274 (A/G). The CASP3 gene polymorphism rs113420705 led to an increase in IL-6.
Conclusion: These findings enhance our understanding of pediatric COVID-19 and may hold promise for developing targeted interventions and providing a personalized medical approach for each patient.
Keywords: COVID-19; children; cytokine; genetic polymorphism; immune response.
© 2025 Kozak, Pavlyshyn, Kamyshnyi, Shevchuk, Korda and Vari.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
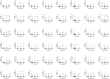
 —a significant difference in two groups comparison (adjusted for multiple testing).
—a significant difference in two groups comparison (adjusted for multiple testing).  —a significant difference in two groups comparison (unadjusted for multiple testing).
—a significant difference in two groups comparison (unadjusted for multiple testing).
 —a significant difference in two groups comparison.
—a significant difference in two groups comparison.References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

