Mpox virus poxin-schlafen fusion protein suppresses innate antiviral response by sequestering STAT2
- PMID: 40066622
- PMCID: PMC11921170
- DOI: 10.1080/22221751.2025.2477639
Mpox virus poxin-schlafen fusion protein suppresses innate antiviral response by sequestering STAT2
Abstract
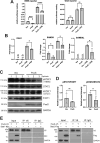
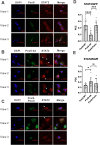
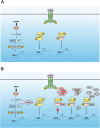
Mpox virus (MPXV) has to establish efficient interferon (IFN) antagonism for effective replication. MPXV-encoded IFN antagonists have not been fully elucidated. In this study, the IFN antagonism of poxin-schlafen (PoxS) fusion gene of MPXV was characterized. MPXV PoxS was capable of decreasing cGAS-produced 2'3'-cGAMP, like its ortholog poxin of vaccinia virus, which is the first known cytosolic nuclease that hydrolyses the 3'-5' bond of 2'3'-cyclic GMP-AMP (cGAMP). However, MPXV PoxS did not suppress cGAS-STING-mediated type I IFN production. Instead, MPXV PoxS antagonized basal and type I IFN-induced expression of IFN-stimulated genes such as OAS1, SAMD9, SAMD9L, ISG15, ISG56 and IFIT3. Consistently, MPXV PoxS inhibited both basal and type I IFN-stimulated activity of interferon-stimulated response elements, but did not affect activation of IFN-γ-activated sites. Mechanistically, MPXV PoxS interacted with STAT2 and sequestered it in the cytoplasm. Both the viral schlafen fusion and the active site of 2'3'-cGAMP nuclease were required for STAT2 sequestration and consequent suppression of IFN-stimulated gene expression. MPXV PoxS conferred resistance to the suppression of MPXV replication by type I IFN. Taken together, our findings suggested that MPXV PoxS counteracts host antiviral response by sequestering STAT2 to circumvent basal and type I IFN-induced expression of antiviral genes.
Keywords: 2′3′-cGAMP hydrolase; STAT2; STING; cGAS; innate antiviral response; monkeypox virus; mpox virus; poxin.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
-
- Masirika LM, Udahemuka JC, Schuele L, et al. . Ongoing mpox outbreak in Kamituga, South Kivu province, associated with monkeypox virus of a novel clade I sub-lineage, Democratic Republic of the Congo, 2024. Euro Surveill. 2024;29:2400106. doi:10.2807/1560-7917.ES.2024.29.11.2400106 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
