The atypical KRASQ22K mutation directs TGF-β response towards partial epithelial-to-mesenchymal transition in patient-derived colorectal cancer tumoroids
- PMID: 40066744
- PMCID: PMC12330932
- DOI: 10.1002/1878-0261.70014
The atypical KRASQ22K mutation directs TGF-β response towards partial epithelial-to-mesenchymal transition in patient-derived colorectal cancer tumoroids
Abstract
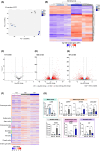
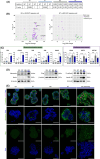
Transforming growth factor beta (TGF-β) exhibits complex and context-dependent cellular responses. While it mostly induces tumor-suppressive effects in early stages of tumorigenesis, tumor-promoting properties are evident in advanced disease. This TGF-β duality is still not fully understood, and whether TGF-β supports invasion and metastasis by influencing cancer cells directly, or rather through the stromal tumor compartment, remains a matter of debate. Here, we utilized a library of colorectal cancer (CRC) patient-derived tumoroids (PDTs), representing a spectrum of tumor stages, to study cancer cell-specific responses to TGF-β. Using conditions allowing for the differentiation of PDTs, we observed TGF-β-induced tumor-suppressive effects in early-stage tumoroids, whereas more advanced tumoroids were less sensitive to the treatment. Notably, one tumoroid line harboring an atypical KRASQ22K mutation underwent partial epithelial-to-mesenchymal transition (EMT), which was associated with morphological changes and increased invasiveness. On a molecular level, this was accompanied by elevated expression of mesenchymal genes, as well as deregulation of pathways associated with matrix remodeling and cell adhesion. Our results suggest that tumor cell-intrinsic responses to TGF-β are critical in determining its tumor-suppressive or tumor-promoting effects.
Keywords: EMT; KRAS mutations; TGF‐β; colorectal cancer; organoids; patient‐derived tumoroids.
© 2025 The Author(s). Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Ihara S, Hirata Y, Koike K. TGF‐β in inflammatory bowel disease: a key regulator of immune cells, epithelium, and the intestinal microbiota. J Gastroenterol. 2017;52(7):777–787. - PubMed
-
- Calon A, Lonardo E, Berenguer‐Llergo A, Espinet E, Hernando‐Momblona X, Iglesias M, et al. Stromal gene expression defines poor‐prognosis subtypes in colorectal cancer. Nat Genet. 2015;47(4):320–329. - PubMed
-
- Tauriello DVF, Sancho E, Batlle E. Overcoming TGFβ‐mediated immune evasion in cancer. Nat Rev Cancer. 2022;22(1):25–44. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

