Eukaryotic initiation factors eIF4F and eIF4B promote translation termination upon closed-loop formation
- PMID: 40066881
- PMCID: PMC11894530
- DOI: 10.1093/nar/gkaf161
Eukaryotic initiation factors eIF4F and eIF4B promote translation termination upon closed-loop formation
Abstract
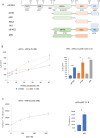
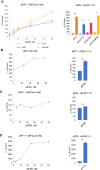
Eukaryotic translation initiation factor 4F (eIF4F), comprising subunits eIF4G, eIF4E, and eIF4A, plays a pivotal role in the 48S preinitiation complex assembly and ribosomal scanning. Additionally, eIF4B enhances the helicase activity of eIF4A. eIF4F also interacts with poly (A)-binding protein (PABP) bound to the poly (A) tail of messenger RNA (mRNA), thereby forming a closed-loop structure. PABP, in turn, interacts with eukaryotic release factor 3 (eRF3), stimulating translation termination. Here, we employed a reconstituted mammalian system to directly demonstrate that eIF4F potently enhances translation termination. Specifically, eIF4A and eIF4B promote the loading of eRF1 into the A site of the ribosome, while eIF4G1 stimulates the GTPase activity of eRF3 and facilitates the dissociation of release factors following peptide release. We also identified MIF4G as the minimal domain required for this activity and showed that eIF4G2/DAP5 can also promote termination. Our findings provide compelling evidence that the closed-loop mRNA structure facilitates translation termination, with PABP and eIF4F directly involved in this process.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

