Function of cytochrome P450 CYP72A1182 in metabolic herbicide resistance evolution in Amaranthus palmeri populations
- PMID: 40067326
- PMCID: PMC12223498
- DOI: 10.1093/jxb/eraf114
Function of cytochrome P450 CYP72A1182 in metabolic herbicide resistance evolution in Amaranthus palmeri populations
Abstract
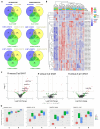
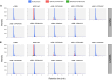
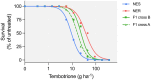
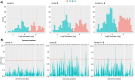
Evolution of metabolic herbicide resistance is a major issue for weed management. Few genes and regulatory mechanisms have been identified, particularly in dicotyledonous weed species. We identified putative causal genes and regulatory mechanism for tembotrione resistance in Palmer amaranth (Amaranthus palmeri). Cytochrome P450 candidate genes were identified through RNA-seq analysis. We validated their functions using heterologous expression in Saccharomyces cerevisae. Promoters of the candidate P450 genes were analysed. We performed QTL mapping to identify genomic regions associated with resistance. CYP72A1182 metabolized tembotrione in a heterologous system. The CYP72A1182 gene had increased expression in other A. palmeri populations resistant to multiple herbicides, including tembotrione. Resistant plants exhibited polymorphisms in the promoter of CYP72A1182. We identified quantitative trait loci linked to herbicide resistance, including one on chromosome 4 approximately 3 Mb away from CYP72A1182. CYP72A1182 is likely involved in tembotrione resistance in A. palmeri. Increased expression of this gene could be due to cis-regulation in the promoter, as well as trans-regulation from transcription factors, and further studies are in progress to test this hypothesis. The elucidation of regulatory genes is crucial for developing innovative weed management approaches and target-based novel herbicide molecules.
Keywords: Gene expression regulation; herbicide resistance; hydroxylation; metabolism; monooxygenase; oxidation.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures

References
-
- Aarthy T, Shyam C, Jugulam M.. 2022. Rapid metabolism and increased expression of CYP81E8 gene confer high level of resistance to tembotrione in a multiple-resistant Palmer amaranth (Amaranthus palmeri S. Watson). Frontiers in Agronomy 4, 1010292.
-
- Andersen CL, Jensen JL, Ørntoft TF.. 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research 64, 5245–5250. - PubMed
-
- Bailey TL, Elkan C.. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings. International Conference on Intelligent Systems for Molecular Biology 2, 28–36. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

