Meta genetic analysis of melon sweetness
- PMID: 40067361
- PMCID: PMC11897113
- DOI: 10.1007/s00122-025-04863-6
Meta genetic analysis of melon sweetness
Abstract
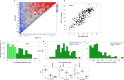
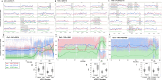
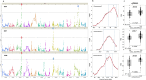
Through meta-genetic analysis of Cucumis melo sweetness, we expand the description of the complex genetic architecture of this trait. Integration of extensive new results with published QTL data provides an outline towards construction of a melon sweetness pan-QTLome. An ultimate objective in crop genetics is describing the complete repertoire of genes and alleles that shape the phenotypic variation of a quantitative trait within a species. Flesh sweetness is a primary determinant of fruit quality and consumer acceptance of melons. Cucumis melo is a diverse species that, among other traits, displays extensive variation in total soluble solids (TSS) content in fruit flesh, ranging from 20 Brix in non-sweet to 180 Brix in sweet accessions. We present here meta-genetic analysis of TSS and sugar variation in melon, using six different populations and fruit measurements collected from more than 30,000 open-field and greenhouse-grown plants, integrated with 15 published melon sweetness-related quantitative trait loci (QTL) studies. Starting with characterization of sugar composition variation across 180 diverse accessions that represent 3 subspecies and 12 of their cultivar-groups, we mapped TSS and sugar QTLs, and confirmed that sucrose accumulation is the key variable explaining TSS variation. All modes-of-inheritance for TSS were displayed by multi-season analysis of a broad half-diallel population derived from 20 diverse founders, with significant prevalence of the additive component. Through parallel genetic mapping in four advanced bi-parental populations, we identified common as well as unique TSS QTLs in 12 chromosomal regions. We demonstrate the cumulative less-than-additive nature of favorable TSS QTL alleles and the potential of a QTL-stacking approach. Using our broad dataset, we were additionally able to show that TSS variation displays weak genetic correlations with melon fruit size and ripening behavior, supporting effective breeding for sweetness per se. Our integrated analysis, combined with additional layers of published QTL data, broadens the perspective on the complex genetic landscape of melon sweetness and proposes a scheme towards future construction of a crop community-driven melon sweetness pan-QTLome.
Keywords: Cucumis melo; Cucurbitaceae; BSA-Seq; Fruit-quality; GWAS; Genome; Half-diallel; QTL mapping; QTLome; Sugars; Total soluble solids (TSS).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: No conflict of interest declared.
Figures



References
-
- Alseekh S, Scossa F, Wen W et al (2021) Domestication of crop metabolomes: Desired and unintended consequences. Trends Plant Sci 26:650–661. 10.1016/j.tplants.2021.02.005 - PubMed
-
- Andrade IS, Melo CAF de, Nunes GH de S, et al (2021) Phenotypic variability, diversity and genetic-population structure in melon (Cucumis melo L.) associated with total soluble solids. Sci Hortic (Amsterdam) 278:. 10.1016/j.scienta.2020.109844
-
- Andrade IS, de Melo CAF, de Nunes GH, S, et al (2019) Morphoagronomic genetic diversity of Brazilian melon accessions based on fruit traits. Sci Hortic (Amsterdam) 243:514–523. 10.1016/j.scienta.2018.09.006
-
- Anon (2015) Growing access to phenotype data. Nat Genet 47:99. 10.1038/ng.3213 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

