Activin A activation of Smad3 mitigates innate inflammation in mouse models of psoriasis and sepsis
- PMID: 40067393
- PMCID: PMC12043092
- DOI: 10.1172/JCI187063
Activin A activation of Smad3 mitigates innate inflammation in mouse models of psoriasis and sepsis
Abstract
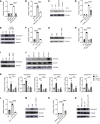
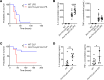
Phosphorylation of Smad3 is a critical mediator of TGF-β signaling, which plays an important role in regulating innate immune responses. However, whether Smad3 activation can be regulated in innate immune cells in TGF-β-independent contexts remains poorly understood. Here, we show that Smad3 is activated through the phosphorylation of its C-terminal residues (pSmad3C) in murine and human macrophages in response to bacterial and viral ligands, and this activation is mediated by activin A in a TGF-β-independent manner. Specifically, infectious ligands, such as LPS, induced secretion of activin A through the transcription factor STAT5 in macrophages, and activin A signaling in turn activated pSmad3C. This activin A/Smad3 axis controlled mitochondrial ATP production and ATP conversion into adenosine by CD73 in macrophages, enforcing an antiinflammatory mechanism. Consequently, mice with a deletion of activin A receptor 1b specifically in macrophages (Acvr1bfl/fl-Lyz2cre) succumbed more to sepsis as a result of uncontrolled inflammation and exhibited exacerbated skin disease in a mouse model of imiquimod-induced psoriasis. Thus, we have revealed a previously unrecognized natural brake to inflammation in macrophages that occurs through the activation of Smad3 in an activin A-dependent manner.
Keywords: Cytokines; Immunology; Inflammation; Innate immunity; Macrophages.
Conflict of interest statement
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

