Genetic subtyping by Whole Exome Sequencing across Diffuse Large B Cell Lymphoma and Plasmablastic Lymphoma
- PMID: 40067847
- PMCID: PMC11896070
- DOI: 10.1371/journal.pone.0318689
Genetic subtyping by Whole Exome Sequencing across Diffuse Large B Cell Lymphoma and Plasmablastic Lymphoma
Abstract
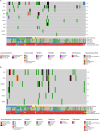
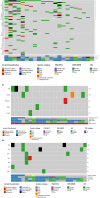
Diffuse Large B-Cell Lymphoma (DLBCL) is a heterogeneous disease characterized by a limited number of molecularly defined subtypes. Recently, genomic-based algorithms have been proposed for the classification of this disease. The whole exome sequencing was conducted on 108 diagnostic samples of diffuse large B-cell lymphoma (DLBCL). Somatic variants, predicted copy number alterations (CNAs), and available fusion data were utilized to classify the cases. Additionally, the enrichment of mutations in the TP53, MYC, and MAPK/ERK pathways was analyzed. Genetic subtypes were identified in approximately 55% of the cases. Cases with a specific genetic subtype exhibited a significantly higher Tumor Mutation Burden compared to molecularly unclassified cases (Mann-Whitney U test, p = 0.024). The prevalence of subtypes varied according to the cell of origin phenotypes. GC-B type DLBCL NOS were classified as EZB (5 cases, 16%), ST2 (5 cases, 16%), and BN2 (1 case, 3%). Four cases (13%) were genetically composite. Three cases of HGBCL/DLBCL double-hit (MYC & BCL2) were classified as EZB-MYC. Forty-three non-GC-B type DLBCL cases were classified as ST2 (5 cases, 11%), BN2 (6 cases, 14%), and MCD (3 cases, 7%). Nine cases were genetically composite (20%). MYC pathway mutations were enriched in cases with EZB and ST2 genetic features, while they were absent in the MCD subtype. TP53 mutations were identified in 11% of the cases. Plasmablastic lymphomas exhibit genetic diversity, with 27% of tumors classified as ST2. Recurrent somatic mutations indicate dysregulation of the JAK/STAT, MAPK/ERK, and tyrosine kinase signaling pathways.
Copyright: © 2025 Pomares et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have the following competing interests to declare: Jaime Perez de Oteyza Speakers bureau: Roche, Incyte, Servier Research funding: Pharmacyclics Angel Ramirez Payer Honoraria: Janssen, BMS, Novartis, Roche, EUSA Pharma, Abbvie, Astra Zeneca, Sanofi, Takeda, Incyte, GSK Consulting/Advisory: Janssen, BMS, Pfizer, Roche, Abbvie, Gilead, Novartis, Takeda; Astra Zeneca, Beigene Eva Gonzalez Barca Consultancy: Johnson & Johnson , Abbvie, Kiowa, Beigene, SOBI, Astra-Zeneca Speaker: Johnson & Johnson, Abbvie, Takeda, Astra-Zeneca, Lilly Travel: Johnson & Johnson, Abbvie, AstraZeneca Sonia Gonzalez Speaker´s Honoraria: Roche, Janssen, EusaPharma, Beigene, Takeda, Incyte, KITE-Pharma Advisory Board: Incyte, SOBI, Roche, Abbvie,Astrazeneca Travel/acommodation: Kite-Pharma, Roche Alejandro Martin Garcia-Sancho Honoraria and or consulting fees: Abbvie, AstraZeneca, BeiGene, BMS, Genmab, Gilead / Kite, GSK, Ideogen, Incyte, Janssen, Kyowa Kirin, Lilly, Miltenyi, Regeneron, Roche, Sobi, Takeda Research support: Gilead / Kite Santiago Montes-Moreno Speaker´s Honoraria: Roche, Janssen, Recordati Rare Diseases There are no patents, products in development or marketed products associated with this research to declare. This does not alter our adherence to PLOS ONE policies on sharing data and materials. The rest of co-authors do not declare any potential conflict of interest related to this paper.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

