Unintegrated HIV-1 DNA recruits cGAS via its histone-binding domain to escape innate immunity
- PMID: 40067888
- PMCID: PMC11929445
- DOI: 10.1073/pnas.2424465122
Unintegrated HIV-1 DNA recruits cGAS via its histone-binding domain to escape innate immunity
Abstract
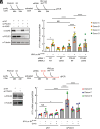
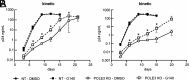
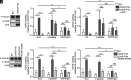
To ensure optimal replication and spread, viruses have evolved countermeasures to evade type 1 IFN-mediated antiviral activity. During the early viral replication cycle steps until uncoating, the HIV-1 core protects viral pathogen associated molecular patterns (viral RNA and reverse transcription products) from recognition by innate immune sensors, including cGAS. However, after capsid uncoating, unintegrated viral DNA (uvDNA) becomes accessible. Here, we show that HIV-1 uses chromatin-mediated cGAS inactivation as a mechanism to protect its uvDNA from innate immune activation.
Keywords: HIV; cGAS; chromatin; innate immunity.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

