Palmitic acid alters enhancers/super-enhancers near inflammatory and efferocytosis-associated genes in human monocytes
- PMID: 40068774
- PMCID: PMC12002881
- DOI: 10.1016/j.jlr.2025.100774
Palmitic acid alters enhancers/super-enhancers near inflammatory and efferocytosis-associated genes in human monocytes
Abstract
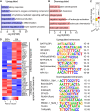
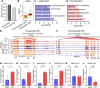
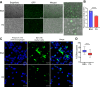
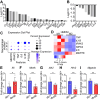
Free fatty acids like palmitic acid (PA) are elevated in obesity and diabetes and dysregulate monocyte and macrophage functions, contributing to enhanced inflammation in these cardiometabolic diseases. Epigenetic mechanisms regulating enhancer functions play key roles in inflammatory gene expression, but their role in PA-induced monocyte/macrophage dysfunction is unknown. We found that PA treatment altered the epigenetic landscape of enhancers and super-enhancers (SEs) in human monocytes. Integration with RNA-seq data revealed that PA-induced enhancers/SEs correlated with PA-increased expression of inflammatory and immune response genes, while PA-inhibited enhancers correlated with downregulation of phagocytosis and efferocytosis genes. These genes were similarly regulated in macrophages from mouse models of diabetes and accelerated atherosclerosis, human atherosclerosis, and infectious agents. PA-regulated enhancers/SEs harbored SNPs associated with diabetes, obesity, and body mass index indicating disease relevance. We verified increased chromatin interactions between PA-regulated enhancers/SEs and inflammatory gene promoters and reduced interactions at efferocytosis genes. PA-induced gene expression was reduced by inhibitors of BRD4, and NF-κB. PA treatment inhibited phagocytosis and efferocytosis in human macrophages. Together, our findings demonstrate that PA-induced enhancer dynamics at key monocyte/macrophage enhancers/SEs regulate inflammatory and immune genes and responses. Targeting these PA-regulated epigenetic changes could provide novel therapeutic opportunities for cardiometabolic disorders.
Keywords: diabetes; dyslipidemias; epigenetic mechanisms; free fatty acids (FFA); genomics; inflammation; macrophage; monocyte; obesity; phagocytosis.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- McGarry J.D. Banting lecture 2001: dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes. 2002;51:7–18. - PubMed
-
- Palomer X., Pizarro-Delgado J., Barroso E., Vazquez-Carrera M. Palmitic and Oleic acid: the Yin and Yang of fatty acids in type 2 diabetes mellitus. Trends Endocrinol. Metab. 2018;29:178–190. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

