Microbiota and metabolite-based prediction tool for colonic polyposis with and without a known genetic driver
- PMID: 40069167
- PMCID: PMC11913376
- DOI: 10.1080/19490976.2025.2474141
Microbiota and metabolite-based prediction tool for colonic polyposis with and without a known genetic driver
Abstract
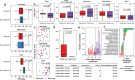
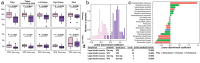
Despite extensive investigations into the microbiome and metabolome changes associated with colon polyps and colorectal cancer (CRC), the microbiome and metabolome profiles of individuals with colonic polyposis, including those with (Gene-pos) and without (Gene-neg) a known genetic driver, remain comparatively unexplored. Using colon biopsies, polyps, and stool from patients with Gene-pos adenomatous polyposis (N = 9), Gene-neg adenomatous polyposis (N = 18), and serrated polyposis syndrome (SPS, N = 11), we demonstrated through 16S rRNA sequencing that the mucosa-associated microbiota in individuals with colonic polyposis is representative of the microbiota associated with small polyps, and that both Gene-pos and SPS cohorts exhibit differential microbiota populations relative to Gene-neg polyposis cohorts. Furthermore, we used these differential microbiota taxa to perform linear discriminant analysis to differentiate Gene-neg subjects from Gene-pos and from SPS subjects with an accuracy of 89% and 93% respectively. Stool metabolites were quantified via 1H NMR, revealing an increase in alanine in SPS subjects relative to non-polyposis subjects, and Partial Least Squares Discriminant Analysis (PLS-DA) analysis indicated that the proportion of leucine to tyrosine in fecal samples may be predictive of SPS. Use of these microbial and metabolomic signatures may allow for better diagnostric and risk-stratification tools for colonic polyposis patients and their families as well as promote development of microbiome-targeted approaches for polyp prevention.
Keywords: Polyposis; alanine; biomarker; colonic adenomatous polyposis of unknown etiology; leucine; metabolome; microbiome; serrated polyposis syndrome; tyrosine.
Conflict of interest statement
No potential conflict of interest was reported by the author(s). All authors reviewed and approved the final manuscript
Figures


References
-
- Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, Hacker-Prietz A, Rabizadeh S, Woster PM, Sears CL, et al. Polyamine catabolism contributes to enterotoxigenic bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci USA. 2011;108(37):15354–15359. doi:10.1073/pnas.1010203108. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
