The DEAD-box helicase eIF4A1/2 acts as RNA chaperone during mitotic exit enabling chromatin decondensation
- PMID: 40069174
- PMCID: PMC11897408
- DOI: 10.1038/s41467-025-57592-1
The DEAD-box helicase eIF4A1/2 acts as RNA chaperone during mitotic exit enabling chromatin decondensation
Abstract
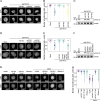
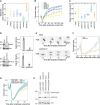
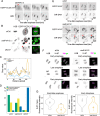
During mitosis, chromosomes condense and decondense to segregate faithfully and undamaged. The exact molecular mechanisms are not well understood. We identify the DEAD-box helicase eIF4A1/2 as a critical factor in this process. In a cell-free condensation assay eIF4A1/2 is crucial for this process, relying on its RNA-binding ability but not its ATPase activity. Reducing eIF4A1/2 levels in cells consistently slows down chromatin decondensation during nuclear reformation. Conversely, increasing eIF4A1/2 concentration on mitotic chromosomes accelerates their decondensation. The absence of eIF4A1/2 affects the perichromatin layer, which surrounds the chromosomes during mitosis and consists of RNA and mainly nucleolar proteins. In vitro, eIF4A1/2 acts as an RNA chaperone, dissociating biomolecular condensates of RNA and perichromatin proteins. During mitosis, the chaperone activity of eIF4A1/2 is required to regulate the composition and fluidity of the perichromatin layer, which is crucial for the dynamic reorganization of chromatin as cells exit mitosis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Nigg, E. A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol.2, 21–32 (2001). - PubMed
-
- Walczak, C. E. & Heald, R. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol.265, 111–158 (2008). - PubMed
-
- Antonin, W. & Neumann, H. Chromosome condensation and decondensation during mitosis. Curr. Opin. Cell Biol.40, 15–22 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

