Ex.50.T aptamer impairs tumor-stroma cross-talk in breast cancer by targeting gremlin-1
- PMID: 40069570
- PMCID: PMC11897156
- DOI: 10.1038/s41420-025-02363-6
Ex.50.T aptamer impairs tumor-stroma cross-talk in breast cancer by targeting gremlin-1
Abstract
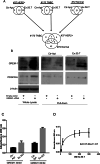
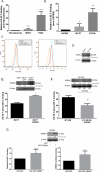
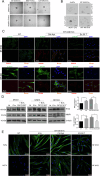
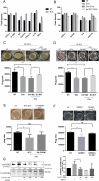
The tumor microenvironment profoundly influences tumor complexity, particularly in breast cancer, where cancer-associated fibroblasts play pivotal roles in tumor progression and therapy resistance. Extracellular vesicles are involved in mediating communication within the TME, specifically highlighting their role in promoting the transformation of normal fibroblasts into cancer-associated fibroblasts. Recently, we identified an RNA aptamer, namely ex.50.T, that binds with remarkable affinity to extracellular vesicles shed from triple-negative breast cancer cells. Here, through in vitro assays and computational analyses, we demonstrate that the binding of ex.50.T to extracellular vesicles and parental breast cancer cells is mediated by recognition of gremlin-1 (GREM1), a bone morphogenic protein antagonist implicated in breast cancer aggressiveness and metastasis. Functionally, we uncover the role of ex.50.T as an innovative therapeutic agent in the process of tumor microenvironment re-modeling, impeding GREM1 signaling, blocking triple-negative breast cancer extracellular vesicles internalization in recipient cells, and counteracting the transformation of normal fibroblasts into cancer-associated fibroblasts. Altogether, our findings highlight ex.50.T as a novel therapeutical avenue for breast cancer and potentially other GREM1-dependent malignancies, offering insights into disrupting TME dynamics and enhancing cancer treatment strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: GC, CQ, CLE, and FI disclose to be inventors of the following patent: ANTICANCER APTAMERS AND USES THEREOF PCT/EP2021/066448. Ethics approval and consent to participate: Samples were collected according to the Declaration of Helsinki, and each subject signed an informed consent form before participating in the study. The study was approved by the Research Ethics Committee of the AUO-University of Naples Federico II no. 119/15ES1.
Figures

References
LinkOut - more resources
Full Text Sources

