Circulating tumor DNA in lymphoma: technologies and applications
- PMID: 40069858
- PMCID: PMC11900646
- DOI: 10.1186/s13045-025-01673-7
Circulating tumor DNA in lymphoma: technologies and applications
Abstract
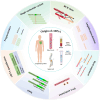
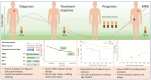
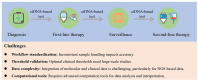
Lymphoma, a malignant tumor derived from lymphocytes and lymphoid tissues, presents with complex and heterogeneous clinical manifestations, requiring accurate patient classification for appropriate treatment. While invasive pathological examination of lymph nodes or lymphoid tissue remains the gold standard for lymphoma diagnosis, its utility is limited in cases of deep-seated tumors such as intraperitoneal and central nervous system lymphomas. In addition, biopsy procedures carry an inherent risk of complications. Computed tomography (CT) and positron emission tomography/computed tomography (PET/CT) imaging are essential for treatment assessment and monitoring, but lack the ability to detect early clonal evolution and minimal residual disease (MRD). Liquid biopsy-based analysis of circulating tumor DNA (ctDNA) offers a non-invasive alternative that allows for repeated sampling and overcomes the limitations of spatial heterogeneity and invasive biopsies. ctDNA provides genetic and epigenetic insights into lymphoma and serves as a dynamic, quantifiable biomarker for diagnosis, risk stratification, and treatment response. This review comprehensively summarizes common genetic variations in lymphoma and systematically evaluates ctDNA detection technologies, including PCR-based assays and next-generation sequencing (NGS). Applications of ctDNA detection in noninvasive genotyping, risk stratification, therapeutic response monitoring, and MRD detection are discussed across various lymphoma subtypes, including diffuse large B-cell lymphoma, Hodgkin lymphoma, follicular lymphoma, and T-cell lymphoma. By integrating recent research findings, the review highlights the role of ctDNA profiling in advancing precision medicine, enabling personalized therapeutic strategies, and improving clinical outcomes in lymphoma.
Keywords: Circulating tumor DNA; Liquid biopsy; Lymphoma; Minimal residue disease; Next-generation sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Global Burden of Disease, Cancer C, Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R, Allen C, Alsharif U, Alvis-Guzman N, Amini E, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: a systematic analysis for the global burden of disease study. JAMA Oncol. 2018;4(11):1553–68. - PMC - PubMed
-
- Chaudhari K, Rizvi S, Syed BA. Non-hodgkin lymphoma therapy landscape. Nat Rev Drug Discov. 2019;18(9):663–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

