Genome-wide characterization and expression profiling of the TGA gene family in sweet orange (Citrus sinensis) reveal CsTGA7 responses to multiple phytohormones and abiotic stresses
- PMID: 40070708
- PMCID: PMC11893830
- DOI: 10.3389/fpls.2025.1530242
Genome-wide characterization and expression profiling of the TGA gene family in sweet orange (Citrus sinensis) reveal CsTGA7 responses to multiple phytohormones and abiotic stresses
Abstract
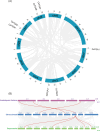
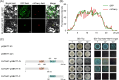
Citrus is widely recognized as one of the most economically important fruit crops worldwide. However, citrus growth is frequently hindered by external environmental stresses, which severely limit its development and yield. The TGA (TGACG motif-binding factor) transcription factors (TFs) are members of the bZIP family and play essential roles in plant defense responses and organ development. Nevertheless, the systematic identification and functional analysis of the TGA family in citrus remains unreported. In this study, genome-wide analysis identified a total of seven CsTGA TFs in Citrus sinensis, which were classified into five subgroups. Phylogenetic and syntenic analysis revealed that the CsTGA genes are highly conserved, with no tandem or segmental duplication events among family members. Promoter sequence analysis identified numerous cis-acting elements associated with transcriptional regulation, phytohormone response, and environmental adaptation in the promoters of CsTGA genes. The expression patterns under five phytohormones and three abiotic stresses demonstrated significant responses of multiple CsTGA genes under various forms of adversity. Among all tested treatments, CsTGA7 showed the most robust response to multiple stresses. Tissue-specific expression pattern analysis revealed potential functional biases among CsTGA genes. In-depth analysis showed that CsTGA7 localized in the nucleus and possessed transcriptional activation activity, consistent with the typical characteristic of transcriptional regulators. In summary, our research systematically investigated the genomic signature of the TGA family in C. sinensis and unearthed CsTGA7 with potential functions in phytohormone signaling transduction and abiotic stress responses. Our study establishes a basis for further exploration of the function of CsTGA genes under abiotic stress.
Keywords: Citrus sinensis; TGA transcription factor; abiotic stress; expression pattern; genome-wide identification.
Copyright © 2025 Wang, Ma, Qiu, Long and Dai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Genome-wide identification of the GRF family in sweet orange (Citrus sinensis) and functional analysis of the CsGRF04 in response to multiple abiotic stresses.BMC Genomics. 2024 Jan 6;25(1):37. doi: 10.1186/s12864-023-09952-8. BMC Genomics. 2024. PMID: 38184538 Free PMC article.
-
Genome-wide identification, characterization, and expression profile ofNBS-LRRgene family in sweet orange (Citrussinensis).Gene. 2023 Feb 20;854:147117. doi: 10.1016/j.gene.2022.147117. Epub 2022 Dec 13. Gene. 2023. PMID: 36526123
-
Genome-wide identification of the trehalose-6-phosphate synthase gene family in sweet orange (Citrus sinensis) and expression analysis in response to phytohormones and abiotic stresses.PeerJ. 2022 Sep 9;10:e13934. doi: 10.7717/peerj.13934. eCollection 2022. PeerJ. 2022. PMID: 36105645 Free PMC article.
-
Genome-Wide Identification, Characterization, and Expression Analysis of the Copper-Containing Amine Oxidase Gene Family in Mangrove Kandelia obovata.Int J Mol Sci. 2023 Dec 9;24(24):17312. doi: 10.3390/ijms242417312. Int J Mol Sci. 2023. PMID: 38139139 Free PMC article. Review.
-
A comprehensive review of TGA transcription factors in plant growth, stress responses, and beyond.Int J Biol Macromol. 2024 Feb;258(Pt 1):128880. doi: 10.1016/j.ijbiomac.2023.128880. Epub 2023 Dec 21. Int J Biol Macromol. 2024. PMID: 38141713 Review.
Cited by
-
Genome-Wide Identification of the GRAS Transcription Factor Family in Sweet Orange and the Regulation of Salt Stress-Enhanced Plant Salt Tolerance in Sweet Orange by CsGRAS15 and CsGRAS27.Biomolecules. 2025 Jun 29;15(7):946. doi: 10.3390/biom15070946. Biomolecules. 2025. PMID: 40723818 Free PMC article.
References
LinkOut - more resources
Full Text Sources

