Vaginal Prevotella timonensis Bacteria Enhance HIV-1 Uptake and Differentially Affect Transmission by Distinct Primary Dendritic Cell Subsets
- PMID: 40071689
- PMCID: PMC11898549
- DOI: 10.1002/eji.202451192
Vaginal Prevotella timonensis Bacteria Enhance HIV-1 Uptake and Differentially Affect Transmission by Distinct Primary Dendritic Cell Subsets
Abstract
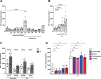
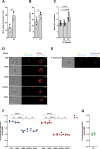
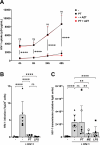
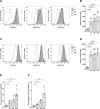
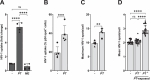
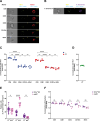
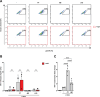
Young females are at high risk of acquiring HIV-1 infections and an imbalance in the vaginal microbiome enhances susceptibility to HIV-1 infection. More insights into the underlying mechanisms could open up new strategies to prevent HIV-1 acquisition and dissemination. Here, we investigated the effect of anaerobic bacteria associated with bacterial vaginosis (BV) on HIV-1 transmission by two distinct dendritic cell (DC) subsets, that is, inflammatory monocyte-derived DCs (moDCs) and primary CD1c+ DCs. Notably, in contrast to other BV-associated microbiota, Prevotella timonensis enhanced uptake of HIV-1 by both moDCs and CD1c+ DCs and the increased uptake was independent of cellular HIV-1 (co-)receptors. Imaging flow cytometry analyses showed that HIV-1 did not co-localise with P. timonensis but was internalized into tetraspanin-positive compartments known to be involved in HIV-1 transmission. P. timonensis bacteria enhanced HIV-1 transmission by CD1c+ DCs, but not by moDCs, and the enhanced transmission was independent of viral infection. Our study strongly suggests that mucosal DC subsets have distinct functions in BV-associated HIV-1 susceptibility, and underscores the importance of early diagnosis and targeted treatment of vaginal dysbiosis to reduce the risk of HIV-1 acquisition.
Keywords: HIV‐1 susceptibility; Prevotella timonensis; dendritic cells (DCs); primary CD1c+ DCs; transmission; vaginal dysbiosis; vaginal microbiome; viral uptake.
© 2025 The Author(s). European Journal of Immunology published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- UNAIDS . UNAIDS Global AIDS update 2022. 2022.
-
- UNAIDS . WE'VE GOT THE POWER Women, Adolescent Girls and the HIV Response. 2020.
-
- Klatt N. R., Cheu R., Birse K., et al., “Vaginal Bacteria Modify HIV Tenofovir Microbicide Efficacy in African Women,” Science 356, no. 6341 (2017): 938–945. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

