Germline variants in CDKN2A wild-type melanoma prone families
- PMID: 40072281
- PMCID: PMC12077288
- DOI: 10.1002/1878-0261.70020
Germline variants in CDKN2A wild-type melanoma prone families
Abstract
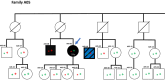
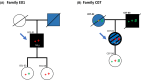
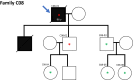
Germline pathogenic variants in CDKN2A are well established as an underlying cause of familial malignant melanoma. While pathogenic variants in other genes have also been linked to melanoma, most familial cases remain unexplained. We assessed pathogenic germline variants in 360 cancer-related genes in 56 Norwegian melanoma-prone families. The index cases were selected based on familial history of melanoma and/or multiple primary melanomas, along with previous negative tests for pathogenic CDKN2A variants. We found 6 out of 56 index individuals to carry germline pathogenic or likely pathogenic variants in BRCA2, MRE11, ATM, MSH2, CHEK2, and AR. One family member with melanoma (not index case) carried a pathogenic variant in MAP3K6. In addition, we found a high fraction of variants previously considered benign and/or as variants of uncertain significance in xeroderma pigmentosum-related genes. In particular, XPCL48F was found in 8 indexes; thus, the allele fraction (0.07) was significantly higher than in comparable healthy populations (0.02-0.03; P-values from 0.007 to 0.014). In conclusion, we found that several melanoma-prone families have pathogenic variants in genes not usually linked to melanoma.
Keywords: cancer risk; germline variants; inheritance; melanoma.
© 2025 The Author(s). Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
Figures


References
-
- Florell SR, Boucher KM, Garibotti G, Astle J, Kerber R, Mineau G, et al. Population‐based analysis of prognostic factors and survival in familial melanoma. J Clin Oncol. 2005;23:7168–7177. - PubMed
-
- Ribeiro Moura Brasil Arnaut J, Dos Santos Guimaraes I, Evangelista Dos Santos AC, de Moraes Lino da Silva F, Machado JR, de Melo AC. Molecular landscape of hereditary melanoma. Crit Rev Oncol Hematol. 2021;164:103425. - PubMed
-
- Helsing P, Nyrnoen DA, Ariansen S, Steine SJ, Maehle L, Aamdal S, et al. Population‐based prevalence of CDKN2A and CDK4 mutations in patients with multiple primary melanomas. Gene Chrom Cancer. 2008;47:175–184. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

