Polygenic Scores of Cardiometabolic Risk Factors in American Indian Adults
- PMID: 40072435
- PMCID: PMC11904716
- DOI: 10.1001/jamanetworkopen.2025.0535
Polygenic Scores of Cardiometabolic Risk Factors in American Indian Adults
Abstract
Importance: Numerous efforts have been made to include diverse populations in genetic studies, but American Indian populations are still severely underrepresented. Polygenic scores derived from genetic data have been proposed in clinical care, but how polygenic scores perform in American Indian individuals and whether they can predict disease risk in this population remains unknown.
Objective: To study the performance of polygenic scores for cardiometabolic risk factors of lipid traits and C-reactive protein in American Indian adults and to determine whether such scores are helpful in clinical prediction for cardiometabolic diseases.
Design, setting, and participants: The Strong Heart Study (SHS) is a large American Indian cohort recruited from 1989 to 1991, with ongoing follow-up (phase VII). In this genetic association study, data from SHS American Indian participants were used in addition to data from 2 large-scale, external, ancestry-mismatched genome-wide association studies (GWASs; 450 865 individuals from a European GWAS and 33 096 individuals from a multi-ancestry GWAS) and 1 small-scale internal ancestry-matched American Indian GWAS (2000 individuals). Analyses were conducted from February 2023 to August 2024.
Exposure: Genetic risk score for cardiometabolic disease risk factors from 6 traits including 5 lipids (apolipoprotein A, apolipoprotein B, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, and triglycerides), and an inflammatory biomarker (C-reactive protein [CRP]).
Main outcomes and measures: Data from SHS participants and the 2 GWASs were used to construct 8 polygenic scores. The association of polygenic scores with cardiometabolic disease was assessed using 2-sided z tests and 1-sided likelihood ratio tests.
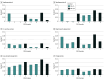
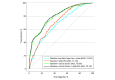
Results: In the 3157 SHS participants (mean [SD] age, 56.44 [8.12] years; 1845 female [58.4%]), a large European-based polygenic score had the most robust performance (mean [SD] R2 = 5.0% [1.7%]), but adding a small-scale ancestry-matched GWAS using American Indian data helped improve polygenic score prediction for 5 of 6 traits (all but CRP; mean [SD] R2, 7.6% [3.2%]). Lipid polygenic scores developed in American Indian individuals improved prediction of diabetes compared with baseline clinical risk factors (area under the curve for absolute improvement, 0.86%; 95% CI, 0.78%-0.93%; likelihood ratio test P = 3.8 × 10-3).
Conclusions and relevance: In this genetic association study of lipids and CRP among American Indian individuals, polygenic scores of lipid traits were found to improve prediction of diabetes when added to clinical risk factors, although the magnitude of improvement was small. The transferability of polygenic scores derived from other populations is still a concern, with implications for the advancement of precision medicine and the potential of perpetuating health disparities, particularly in this underrepresented population.
Conflict of interest statement
Figures
Comment in
- doi: 10.1001/jamanetworkopen.2025.0545
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 MD012765/MD/NIMHD NIH HHS/United States
- R01 HL163972/HL/NHLBI NIH HHS/United States
- R01 HG013163/HG/NHGRI NIH HHS/United States
- R01 DK117445/DK/NIDDK NIH HHS/United States
- U01 HG011720/HG/NHGRI NIH HHS/United States
- R01 HL109315/HL/NHLBI NIH HHS/United States
- R01 HL109301/HL/NHLBI NIH HHS/United States
- R01 HL109284/HL/NHLBI NIH HHS/United States
- R01 HL109282/HL/NHLBI NIH HHS/United States
- R01 HL109319/HL/NHLBI NIH HHS/United States
- U01 HL041642/HL/NHLBI NIH HHS/United States
- U01 HL041652/HL/NHLBI NIH HHS/United States
- U01 HL041654/HL/NHLBI NIH HHS/United States
- U01 HL065520/HL/NHLBI NIH HHS/United States
- U01 HL065521/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

