Carbapenem-Resistant, Virulence Plasmid-Harboring Klebsiella pneumoniae, United States
- PMID: 40072602
- PMCID: PMC11950267
- DOI: 10.3201/eid3104.241396
Carbapenem-Resistant, Virulence Plasmid-Harboring Klebsiella pneumoniae, United States
Abstract
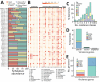
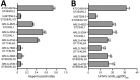
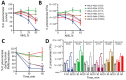
Carbapenem-resistant and virulence plasmid-harboring Klebsiella pneumoniae (pVir-CRKP) has emerged and spread globally, yet clinical investigations from the United States remain limited. We conducted a genomic analysis of 884 unique carbapenem-resistant K. pneumoniae isolates from a multicenter US cohort and identified 6 pVir-CRKP isolates, including 2 sequence type (ST) 23, 2 ST893, and 2 ST11 isolates. Patients infected with pVir-CRKP experienced high Pitt bacteremia scores and a 33% 30-day mortality rate. The pVir-CRKP isolates exhibited significant sequence variation in virulence genes and plasmids, along with differences in mucoviscosity, capsule production, survival in normal human serum, resistance to killing by human polymorphonuclear neutrophils, and in vivo pathogenicity. Phylogenetic analyses showed that most pVir-CRKP isolates were genetically similar to strains reported from other global regions. The emergence of pVir-CRKP with higher virulence potential and carbapenem resistance in the United States than the predominant carbapenem-resistant K. pneumoniae clone underscores the need for active global surveillance.
Keywords: Klebsiella pneumoniae; United States; antimicrobial resistance; bacteria; carbapenem resistance; clinical and genomic characterization; pVir-CRKP; virulence; virulence plasmid.
Figures


References
-
- World Health Organization. Antimicrobial resistance, hypervirulent Klebsiella pneumoniae—global situation. 2024. [cited 2024 Sep 20] https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON527
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

