FlyVISTA, an integrated machine learning platform for deep phenotyping of sleep in Drosophila
- PMID: 40073129
- PMCID: PMC11900856
- DOI: 10.1126/sciadv.adq8131
FlyVISTA, an integrated machine learning platform for deep phenotyping of sleep in Drosophila
Abstract
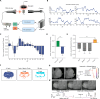
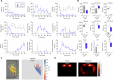
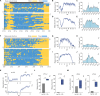
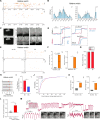
There is great interest in using genetically tractable organisms such as Drosophila to gain insights into the regulation and function of sleep. However, sleep phenotyping in Drosophila has largely relied on simple measures of locomotor inactivity. Here, we present FlyVISTA, a machine learning platform to perform deep phenotyping of sleep in flies. This platform comprises a high-resolution closed-loop video imaging system, coupled with a deep learning network to annotate 35 body parts, and a computational pipeline to extract behaviors from high-dimensional data. FlyVISTA reveals the distinct spatiotemporal dynamics of sleep and wake-associated microbehaviors at baseline, following administration of the sleep-inducing drug gaboxadol, and with dorsal fan-shaped body drivers. We identify a microbehavior ("haltere switch") exclusively seen during quiescence that indicates a deeper sleep stage. These results enable the rigorous analysis of sleep in Drosophila and set the stage for computational analyses of microbehaviors in quiescent animals.
Figures




Update of
-
FlyVISTA, an Integrated Machine Learning Platform for Deep Phenotyping of Sleep in Drosophila.bioRxiv [Preprint]. 2024 Jun 25:2023.10.30.564733. doi: 10.1101/2023.10.30.564733. bioRxiv. 2024. Update in: Sci Adv. 2025 Mar 14;11(11):eadq8131. doi: 10.1126/sciadv.adq8131. PMID: 37961473 Free PMC article. Updated. Preprint.
References
-
- Anafi R. C., Kayser M. S., Raizen D. M., Exploring phylogeny to find the function of sleep. Nat. Rev. Neurosci. 20, 109–116 (2019). - PubMed
-
- Iglesias T. L., Boal J. G., Frank M. G., Zeil J., Hanlon R. T., Cyclic nature of the REM sleep-like state in the cuttlefish Sepia officinalis. J. Exp. Biol. 222, jeb4862 (2019). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

