MYC ecDNA promotes intratumour heterogeneity and plasticity in PDAC
- PMID: 40074906
- PMCID: PMC12003172
- DOI: 10.1038/s41586-025-08721-9
MYC ecDNA promotes intratumour heterogeneity and plasticity in PDAC
Abstract
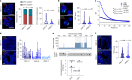
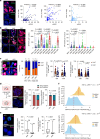
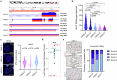
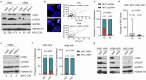
Intratumour heterogeneity and phenotypic plasticity drive tumour progression and therapy resistance1,2. Oncogene dosage variation contributes to cell-state transitions and phenotypic heterogeneity3, thereby providing a substrate for somatic evolution. Nonetheless, the genetic mechanisms underlying phenotypic heterogeneity are still poorly understood. Here we show that extrachromosomal DNA (ecDNA) is a major source of high-level focal amplification in key oncogenes and a major contributor of MYC heterogeneity in pancreatic ductal adenocarcinoma (PDAC). We demonstrate that ecDNAs drive varying levels of MYC dosage, depending on their regulatory landscape, enabling cancer cells to rapidly and reversibly adapt to microenvironmental changes. In the absence of selective pressure, a high ecDNA copy number imposes a substantial fitness cost on PDAC cells. We also show that MYC dosage affects cell morphology and dependence of cancer cells on stromal niche factors. Our work provides a detailed analysis of ecDNAs in PDAC and describes a new genetic mechanism driving MYC heterogeneity in PDAC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures









References
-
- Levan, G., Mandahl, N., Bregula, U., Klein, G. & Levan, A. Double minute chromosomes are not centromeric regions of the host chromosomes. Hereditas83, 83–90 (1976). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

