Proteogenomic Profiling of Treatment-Naïve Metastatic Malignant Melanoma
- PMID: 40075679
- PMCID: PMC11899103
- DOI: 10.3390/cancers17050832
Proteogenomic Profiling of Treatment-Naïve Metastatic Malignant Melanoma
Abstract
Background: Melanoma is a highly heterogeneous disease, and a deeper molecular classification is essential for improving patient stratification and treatment approaches. Here, we describe the histopathology-driven proteogenomic landscape of 142 treatment-naïve metastatic melanoma samples to uncover molecular subtypes and clinically relevant biomarkers.
Methods: We performed an integrative proteogenomic analysis to identify proteomic subtypes, assess the impact of BRAF V600 mutations, and study the molecular profiles and cellular composition of the tumor microenvironment. Clinical and histopathological data were used to support findings related to tissue morphology, disease progression, and patient outcomes.
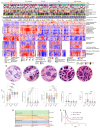
Results: Our analysis revealed five distinct proteomic subtypes that integrate immune and stromal microenvironment components and correlate with clinical and histopathological parameters. We demonstrated that BRAF V600-mutated melanomas exhibit biological heterogeneity, where an oncogene-induced senescence-like phenotype is associated with improved survival. This led to a proposed mortality risk-based stratification that may contribute to more personalized treatment strategies. Furthermore, tumor microenvironment composition strongly correlated with disease progression and patient outcomes, highlighting a histopathological connective tissue-to-tumor ratio assessment as a potential decision-making tool. We identified a melanoma-associated SAAV signature linked to extracellular matrix remodeling and SAAV-derived neoantigens as potential targets for anti-tumor immune responses.
Conclusions: This study provides a comprehensive stratification of metastatic melanoma, integrating proteogenomic insights with histopathological features. The findings may aid in the development of tailored diagnostic and therapeutic strategies, improving patient management and outcomes.
Keywords: BRAF V600E; histopathology; lymph node metastases; melanoma; proteogenomics; proteomics; single amino acid variants; stratification; subtypes; tumor microenvironment.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
Grants and funding
- ÚNKP-22-3-II/National Research, Development and Innovation Fund (Hungary)
- FBKS-2023-22-99/Berta Kamprad Foundation
- STIA-KFI2021/Semmelweis University grant
- OTKA FK138696/Hungarian National Research, Development and Innovation Office
- U24 CA210972/CA/NCI NIH HHS/United States
- OTKA-125509/Hungarian Academy of Sciences
- EFOP-3.6.3-VEKOP-16-2017-00009/Semmelweis 250+ Excellence Ph.D. Scholarship
- FBKS-2024-19 - 604/Berta Kamprad Foundation IKEA
- ÚNKP-21-3- SZTE-102/National Research, Development and Innovation Fund (Hungary)
- U24CA210972/National Cancer Institute (NCI) CPTAC grant
LinkOut - more resources
Full Text Sources
Research Materials

