Spatial Transcriptomics Reveals Novel Mechanisms Involved in Perineural Invasion in Pancreatic Ductal Adenocarcinomas
- PMID: 40075699
- PMCID: PMC11899704
- DOI: 10.3390/cancers17050852
Spatial Transcriptomics Reveals Novel Mechanisms Involved in Perineural Invasion in Pancreatic Ductal Adenocarcinomas
Abstract
Background: Pancreatic ductal adenocarcinoma (PDAC) has a high incidence of perineural invasion (PNI), a pathological feature of the cancer invasion of nerves. PNI is associated with a poor prognosis, local recurrence and cancer pain. It has been suggested that interactions between nerves and the tumor microenvironment (TME) play a role in PDAC tumorigenesis.
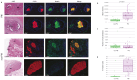
Methods: Here, we used Nanostring GeoMx Digital Spatial Profiler to analyze the whole transcriptome of both cancer and nerve cells in the microenvironment of PNI and non-PNI foci from 13 PDAC patients.
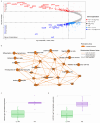
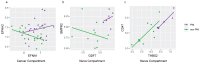
Conclusions: We identified previously reported pathways involved in PNI, including Axonal Guidance and ROBO-SLIT Signaling. Spatial transcriptomics highlighted the role of PNI foci in influencing the immune landscape of the TME and similarities between PNI and nerve injury response. This study revealed that endocannabinoid and polyamine metabolism may contribute to PNI, cancer growth and cancer pain. Key members of these pathways can be targeted, offering potential novel research avenues for exploring new cancer treatment and/or pain management options in PDAC.
Keywords: MGLL; SAT1; nerve; pain; perineural invasion; spatial transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Ceyhan G.O., Bergmann F., Kadihasanoglu M., Altintas B., Demir I.E., Hinz U., Muller M.W., Giese T., Buchler M.W., Giese N.A., et al. Pancreatic neuropathy and neuropathic pain—A comprehensive pathomorphological study of 546 cases. Gastroenterology. 2009;136:177–186. doi: 10.1053/j.gastro.2008.09.029. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

