Deciphering Colorectal Cancer-Hepatocyte Interactions: A Multiomics Platform for Interrogation of Metabolic Crosstalk in the Liver-Tumor Microenvironment
- PMID: 40076609
- PMCID: PMC11900982
- DOI: 10.3390/ijms26051976
Deciphering Colorectal Cancer-Hepatocyte Interactions: A Multiomics Platform for Interrogation of Metabolic Crosstalk in the Liver-Tumor Microenvironment
Abstract
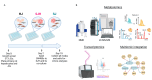
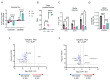
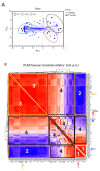
Metabolic reprogramming is a hallmark of cancer, enabling tumor cells to adapt to and exploit their microenvironment for sustained growth. The liver is a common site of metastasis, but the interactions between tumor cells and hepatocytes remain poorly understood. In the context of liver metastasis, these interactions play a crucial role in promoting tumor survival and progression. This study leverages multiomics coverage of the microenvironment via liquid chromatography and high-resolution, high-mass-accuracy mass spectrometry-based untargeted metabolomics, 13C-stable isotope tracing, and RNA sequencing to uncover the metabolic impact of co-localized primary hepatocytes and a colon adenocarcinoma cell line, SW480, using a 2D co-culture model. Metabolic profiling revealed disrupted Warburg metabolism with an 80% decrease in glucose consumption and 94% decrease in lactate production by hepatocyte-SW480 co-cultures relative to SW480 control cultures. Decreased glucose consumption was coupled with alterations in glutamine and ketone body metabolism, suggesting a possible fuel switch upon co-culturing. Further, integrated multiomics analysis indicates that disruptions in metabolic pathways, including nucleoside biosynthesis, amino acids, and TCA cycle, correlate with altered SW480 transcriptional profiles and highlight the importance of redox homeostasis in tumor adaptation. Finally, these findings were replicated in three-dimensional microtissue organoids. Taken together, these studies support a bioinformatic approach to study metabolic crosstalk and discovery of potential therapeutic targets in preclinical models of the tumor microenvironment.
Keywords: cancer metabolism; metabolomics; multiomics; tumor microenvironment.
Conflict of interest statement
P.A.C. has served as an external consultant for Pfizer, Inc., Abbott Laboratories, Janssen Research & Development and Juvenescence. A.C. is now an employee of Solventum.
Figures




Update of
-
Deciphering Colorectal Cancer-Hepatocyte Interactions: A Multiomic Platform for Interrogation of Metabolic Crosstalk in the Liver-Tumor Microenvironment.bioRxiv [Preprint]. 2024 Dec 14:2024.12.06.627264. doi: 10.1101/2024.12.06.627264. bioRxiv. 2024. Update in: Int J Mol Sci. 2025 Feb 25;26(5):1976. doi: 10.3390/ijms26051976. PMID: 39713297 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

