Exploring the Prevalence of SMN1 Duplication and Deletion in Russia and Its Impact on Carrier Screening
- PMID: 40076610
- PMCID: PMC11900099
- DOI: 10.3390/ijms26051984
Exploring the Prevalence of SMN1 Duplication and Deletion in Russia and Its Impact on Carrier Screening
Abstract
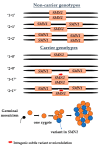
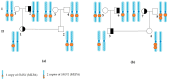
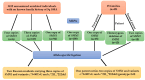
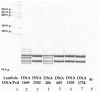
5q spinal muscular atrophy (5q SMA) is one of the most prevalent autosomal recessive disorders worldwide. In 5q SMA, cases of silent carriers have been reported, including SMN1 duplications, intragenic subtle variants, de novo variants, and mosaicism. This study included DNA samples from 3412 unexamined unrelated individuals with no known family history of 5q SMA. In addition, we studied 15 families in which the children had a confirmed diagnosis of 5q SMA caused by a homozygous deletion of exon 7 of SMN1. Each family included one parent who was a carrier of a heterozygous deletion of SMN1, while the other parent had two copies of SMN1. The copy number of SMN1 and SMN2 was detected by MLPA. Two previously reported genetic markers of SMN1 duplication, c.*3+80T>G and c.*211_*212del, were tested in 143 Russian residents with three copies of SMN1 and 15 parents with two copies of SMN1. The frequency of a heterozygous carrier of exon 7 deletion of SMN1 is 1 in 36 individuals (95% CI 33 to 39). The frequency of exon 7 duplication of SMN1 is 1 in 25 individuals (95% CI 20 to 30). Only three individuals of the studied SMN1 duplication carriers were detected to have genetic markers of SMN1 duplication. The study of SMN1 duplication genetic markers (c.*3+80T>G and c.*211_*212del) in Russian residents reveals only 1.9% of SMN1 duplication carriers.
Keywords: 5q SMA; SMN1; c.*211_*212del; c.*3+80T>G; carrier screening; deletion; duplication; genotype (2 + 0); markers; silent carrier; spinal muscular atrophy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Luo M., Liu L., Peter I., Zhu J., Scott S.A., Zhao G., Eversley C., Kornreich R., Desnick R.J., Edelmann L. An Ashkenazi Jewish SMN1 haplotype specific to duplication alleles improves pan-ethnic carrier screening for spinal muscular atrophy. Genet. Med. 2014;16:149–156. doi: 10.1038/gim.2013.84. - DOI - PubMed
-
- Gregg A.R., Aarabi M., Klugman S., Leach N.T., Bashford M.T., Goldwaser T., Chen E., Sparks T.N., Reddi H.V., Rajkovic A., et al. ACMG Professional Practice and Guidelines Committee. Screening for autosomal recessive and X-linked conditions during pregnancy and preconception: A practice resource of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2021;23:1793–1806. doi: 10.1038/s41436-021-01203-z. Erratum in Genet. Med. 2021, 23, 2015. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

