A Novel 6-bp Repeat Unit (6-bp RU) of the 13th Intron Within the Conserved EPAS1 Gene in Plateau Pika Is Capable of Altering Enhancer Activity
- PMID: 40076786
- PMCID: PMC11901085
- DOI: 10.3390/ijms26052163
A Novel 6-bp Repeat Unit (6-bp RU) of the 13th Intron Within the Conserved EPAS1 Gene in Plateau Pika Is Capable of Altering Enhancer Activity
Abstract
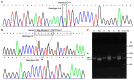
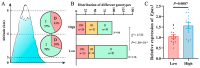
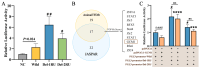
The plateau pika (pl-pika), a resilient mammal of the Qinghai-Tibet Plateau, exhibits remarkable adaptations to extreme conditions. This study delves into mutations within the Endothelial PAS Domain Protein 1 (EPAS1) gene, crucial for high-altitude survival. Surprisingly, a novel 6-bp insertion/deletion (indel) mutation in EPAS1's Intron 13, along with an additional repeat unit downstream, was discovered during PCR amplification. Genetic analysis across altitude gradients revealed a correlation between this indel's frequency and altitude, hinting at its role in altitude adaptation. Fluorescence enzyme assays unveiled enhancer activity within Intron 13, where the deletion of repeat units led to increased activity, indicating potential transcription factor binding. Notably, GCM1 emerged as a candidate transcription factor binding to the indel site, suggesting its involvement in EPAS1 regulation. These findings enrich our comprehension of high-altitude adaptation in plateau pikas, shedding light on the intricate interplay between genetic mutations, transcriptional regulation, and environmental pressures in evolutionary biology.
Keywords: EPAS1 gene; enhancer; indel; plateau pika; transcription factor.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
A highland-adaptation mutation of the Epas1 protein increases its stability and disrupts the circadian clock in the plateau pika.Cell Rep. 2022 May 17;39(7):110816. doi: 10.1016/j.celrep.2022.110816. Cell Rep. 2022. PMID: 35584682
-
Inhibition of inducible nitric oxide synthase expression and nitric oxide production in plateau pika (Ochotona curzoniae) at high altitude on Qinghai-Tibet Plateau.Nitric Oxide. 2014 Apr 30;38:38-44. doi: 10.1016/j.niox.2014.02.009. Epub 2014 Mar 12. Nitric Oxide. 2014. PMID: 24632467
-
Two functional loci in the promoter of EPAS1 gene involved in high-altitude adaptation of Tibetans.Sci Rep. 2014 Dec 12;4:7465. doi: 10.1038/srep07465. Sci Rep. 2014. PMID: 25501874 Free PMC article.
-
Adaptive genetic changes related to haemoglobin concentration in native high-altitude Tibetans.Exp Physiol. 2015 Nov;100(11):1263-8. doi: 10.1113/EP085035. Exp Physiol. 2015. PMID: 26454145 Review.
-
Energy power in mountains: difference in metabolism pattern results in different adaption traits in Tibetans.Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2012 Nov;28(6):488-93. Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2012. PMID: 23581177 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

