Emulating computer models with high-dimensional count output
- PMID: 40078142
- PMCID: PMC11904617
- DOI: 10.1098/rsta.2024.0216
Emulating computer models with high-dimensional count output
Abstract
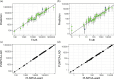
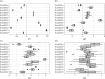
Computer models are used to study the real world, and often contain a large number of uncertain input parameters, produce a large number of outputs, may be expensive to run and need calibrating to real-world observations to be useful for decision-making. Emulators are often used as cheap surrogates for the expensive simulator, trained on a small number of simulations to provide predictions with uncertainty at unseen inputs. In epidemiological applications, for example compartmental or agent-based models for modelling the spread of infectious diseases, the output is usually spatially and temporally indexed, stochastic and consists of counts rather than continuous variables. Here, we consider emulating high-dimensional count output from a complex computer model using a Poisson lognormal PCA (PLNPCA) emulator. We apply the PLNPCA emulator to output fields from a COVID-19 model for England and Wales and compare this to fitting emulators to aggregations of the full output. We show that performance is generally comparable, while the PLNPCA emulator inherits desirable properties, including allowing the full output to be predicted while capturing correlations between outputs, providing high-dimensional samples of counts that are representative of the true model output.This article is part of the theme issue 'Uncertainty quantification for healthcare and biological systems (Part 1)'.
Keywords: Gaussian processes; Poisson lognormal; basis emulation; uncertainty quantification.
Conflict of interest statement
We declare we have no competing interests.
Figures



References
-
- Williamson D, Goldstein M, Allison L, Blaker A, Challenor P, Jackson L, Yamazaki K. 2013. History matching for exploring and reducing climate model parameter space using observations and a large perturbed physics ensemble. Clim. Dyn. 41, 1703–1729. (10.1007/s00382-013-1896-4) - DOI
-
- Salter JM, Williamson DB, Scinocca J, Kharin V. 2019. Uncertainty Quantification for Computer Models With Spatial Output Using Calibration-Optimal Bases. J. Am. Stat. Assoc. 114, 1800–1814. (10.1080/01621459.2018.1514306) - DOI
-
- McNeall D, Robertson E, Wiltshire A. 2024. Constraining the carbon cycle in JULES-ES-1.0. Geosci. Model Dev. 17, 1059–1089. (10.5194/gmd-17-1059-2024) - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

