Riverine antibiotic resistome along an anthropogenic gradient
- PMID: 40078550
- PMCID: PMC11897494
- DOI: 10.3389/fmicb.2025.1516033
Riverine antibiotic resistome along an anthropogenic gradient
Abstract
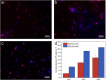
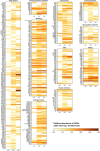
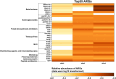
The introduction of antibiotic-resistant bacteria into riverine systems through the discharge of wastewater treatment plant (WWTP) effluent and agricultural waste poses significant health risks. Even when not pathogenic, these bacteria can act as reservoirs for antibiotic resistance genes (ARGs), transferring them to pathogens that infect humans and animals. In this study, we used fluorescence in situ hybridization, qPCR, and metagenomics to investigate how anthropogenic activities affect microbial abundance and the resistome along the Holtemme River, a small river in Germany, from near-pristine to human-impacted sites. Our results showed higher bacterial abundance, a greater absolute and relative abundance of ARGs, and a more diverse ARG profile at the impacted sites. Overall, the ARG profiles at these sites reflected antibiotic usage in Germany, with genes conferring resistance to drug classes such as beta-lactams, aminoglycosides, folate biosynthesis inhibitors, and tetracyclines. There were also variations in the ARG profiles of the impacted sites. Notably, there was a high abundance of the oxacillin resistance gene OXA-4 at the downstream site in the river. In the metagenome assembly, this gene was associated with a contig homologous to small plasmids previously identified in members of the Thiotrichaceae. The likely in-situ host of the putative plasmid was a close relative of Thiolinea (also known as Thiothrix) eikelboomii, a prominent member of WWTP microbiomes worldwide. Our results show that the effluent from WWTPs can introduce bacteria into the environment that act as shuttle systems for clinically relevant ARG.
Keywords: anthropogenic activities; antibiotic resistome; fluorescence in situ hybridization; metagenomic sequencing; riverine system.
Copyright © 2025 Wang, Haenelt, Corrêa, da Rocha, Musat, Zhang, Müller and Musat.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

References
-
- Abia A. L. K., Alisoltani A., Keshri J., Ubomba-Jaswa E. (2018). Metagenomic analysis of the bacterial communities and their functional profiles in water and sediments of the Apies River, South Africa, as a function of land use. Sci. Total Environ. 616–617, 326–334. doi: 10.1016/j.scitotenv.2017.10.322, PMID: - DOI - PubMed
-
- Alneberg J., Bjarnason B. S., de Bruijn I., Schirmer M., Quick J., Ijaz U. Z., et al. . (2013). CONCOCT: Clustering contigs on coverage and composition. arXiv Prepr arXiv, 1–28. Available at: http://arxiv.org/abs/1312.4038 (Accessed February 16, 2025).
-
- Amarasiri M., Sano D., Suzuki S. (2020). Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 50, 2016–2059. doi: 10.1080/10643389.2019.1692611 - DOI
-
- Aoki M., Takemura Y., Kawakami S., Yoochatchaval W., Thao Tran P., Tomioka N., et al. . (2023). Quantitative detection and reduction of potentially pathogenic bacterial groups of Aeromonas, Arcobacter, Klebsiella pneumoniae species complex, and Mycobacterium in wastewater treatment facilities. PLoS One 18, e0291742–e0291725. doi: 10.1371/journal.pone.0291742, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

