Enforcement of stem-cell dormancy by nucleophosmin mutation is a critical determinant of unrestricted self-renewal during myeloid leukemogenesis
- PMID: 40079083
- PMCID: PMC12399960
- DOI: 10.3324/haematol.2024.286577
Enforcement of stem-cell dormancy by nucleophosmin mutation is a critical determinant of unrestricted self-renewal during myeloid leukemogenesis
Abstract
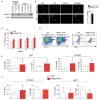
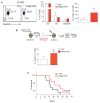
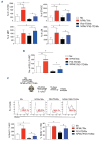
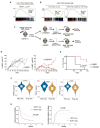
Mutations in the NPM1 gene (NPMc+) and in the FLT3 gene (FLT3-ITD) represent the most frequent co-occurring mutations in acute myeloid leukemia (AML), yet the cellular and molecular mechanisms of their co-operation remain largely unexplored. Using mouse models that faithfully recapitulate human AML, we investigated the impact of these oncogenes on pre-leukemic and leukemic hematopoietic stem cells (HSC), both separately and in combination. While both NPMc+ and Flt3-ITD promote the proliferation of pre-leukemia HSC, only NPMc+ drives extended self-renewal by preventing the depletion of the quiescent HSC pool. Quiescent HSC have a dynamic equilibrium between dormant and active states, which respectively support self-renewal and regenerative hematopoiesis. Transcriptional profiling of these dormant and active states revealed that not only does NPMc+ stimulate the transition from dormancy to activity, but it also reinforces the dormant state, thereby ensuring the replenishment of dormant HSC. Intriguingly, the co-expression of NPMc+ and Flt3-ITD engenders a novel phenotypic state within quiescent HSC, whereby dormancy and activity co-exist within a single cell. We posit that this unique state fuels the in vivo expansion of self-renewing HSC and facilitates the rapid selection of leukemia-initiating cells. Pharmacological inhibition of the dormancy-related TGFβ1 pathway effectively reduces the self-renewal capacity of leukemia stem cells and extends survival in our mouse models. Collectively, these findings demonstrate that enforcement of HSC dormancy is a critical determinant of unrestricted self-renewal during leukemogenesis and, as such, represents a compelling target for the development of novel anti-leukemic therapies.
Figures



References
-
- Mallardo M, Caronno A, Pruneri G, et al. NPMc+ and FLT3_ITD mutations cooperate in inducing acute leukaemia in a novel mouse model. Leukemia. 2013;27(11):2248-2251. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

