Maximizing Immunopeptidomics-Based Bacterial Epitope Discovery by Multiple Search Engines and Rescoring
- PMID: 40080147
- PMCID: PMC11976845
- DOI: 10.1021/acs.jproteome.4c00864
Maximizing Immunopeptidomics-Based Bacterial Epitope Discovery by Multiple Search Engines and Rescoring
Abstract
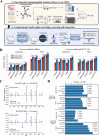
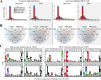
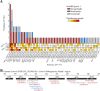
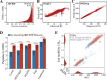
Mass spectrometry-based discovery of bacterial immunopeptides presented by infected cells allows untargeted discovery of bacterial antigens that can serve as vaccine candidates. However, reliable identification of bacterial epitopes is challenged by their extremely low abundance. Here, we describe an optimized bioinformatic framework to enhance the confident identification of bacterial immunopeptides. Immunopeptidomics data of cell cultures infected with Listeria monocytogenes were searched by four different search engines, PEAKS, Comet, Sage and MSFragger, followed by data-driven rescoring with MS2Rescore. Compared with individual search engine results, this integrated workflow boosted immunopeptide identification by an average of 27% and led to the high-confidence detection of 18 additional bacterial peptides (+27%) matching 15 different Listeria proteins (+36%). Despite the strong agreement between the search engines, a small number of spectra (<1%) had ambiguous matches to multiple peptides and were excluded to ensure high-confidence identifications. Finally, we demonstrate our workflow with sensitive timsTOF SCP data acquisition and find that rescoring, now with inclusion of ion mobility features, identifies 76% more peptides compared to Q Exactive HF acquisition. Together, our results demonstrate how integration of multiple search engine results along with data-driven rescoring maximizes immunopeptide identification, boosting the detection of high-confidence bacterial epitopes for vaccine development.
Keywords: Listeria monocytogenes; TIMS2Rescore; immunopeptides; ion mobility; mass spectrometry; search engines.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Murphy K. M.; Weaver C.. Janeway’s Immunobiology, 9th ed.; Garland Science: New York, 2017.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

