The added value of metagenomic next-generation sequencing in central nervous system infections: a systematic review of case reports
- PMID: 40080343
- PMCID: PMC12137511
- DOI: 10.1007/s15010-025-02502-2
The added value of metagenomic next-generation sequencing in central nervous system infections: a systematic review of case reports
Abstract
Background: The diversity of pathogens causing central nervous system (CNS) infections presents a diagnostic challenge. Patient demographics and geographical location affect the likelihood of certain pathogens causing infection. Current diagnostic methods rely on labour-intensive cultivation or targeted detection. Metagenomic next-generation sequencing (mNGS) is a promising tool for detecting pathogens in CNS infections, offering an unbiased approach. To enhance our understanding of patient demographics and the range of pathogens identified through mNGS, we conducted a systematic review of case reports.
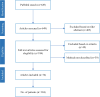
Methods: The PubMed database was searched in March 2024. Case reports on CNS infections and mNGS published from January 2014 through February 2024 were included based on predefined criteria.
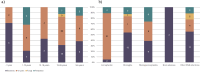
Results: The search yielded 649 articles, of which 76 were included, encompassing 104 patients. Most patients were male (75%), the median age was 31,5 years [0-75] and 28% were immunocompromised. The most common diagnosis was encephalitis (36%), followed by meningitis (23%) and meningoencephalitis (22%). 53 unique pathogens were identified, comprising 27 different viruses, 19 bacteria, 5 parasites, and 2 fungi. Syndromic encephalitis/meningitis panels would only have detected four of the viruses and five of the bacteria. Additionally, 14 of the bacterial species are considered slow-growing or fastidious and could be challenging to detect by culture.
Conclusion: The application of mNGS in diagnosing CNS infections reveals the diversity of pathogens responsible for these severe infections, thereby improving diagnostics and facilitating targeted treatment. While case reports may be subjected to bias, they provide valuable insights into the use of mNGS in this clinical context.
Keywords: Central nervous system infections; Diagnosis; Metagenomic next-generation sequencing; Pathogen indentification.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


References
-
- Granerod J, Ambrose HE, Davies NW, Clewley JP, Walsh AL, Morgan D, et al. Causes of encephalitis and differences in their clinical presentations in England: a multicentre, population-based prospective study. Lancet Infect Dis. 2010;10(12):835–44. 10.1016/s1473-3099(10)70222-x. - PubMed
-
- Solomon T, Michael BD, Smith PE, Sanderson F, Davies NW, Hart IJ, et al. Management of suspected viral encephalitis in adults–Association of British neurologists and British infection association National guidelines. J Infect. 2012;64(4):347–73. 10.1016/j.jinf.2011.11.014. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

