XanthoMoClo─A Robust Modular Cloning Genetic Toolkit for the Genera Xanthobacter and Roseixanthobacter
- PMID: 40080684
- PMCID: PMC12012871
- DOI: 10.1021/acssynbio.4c00806
XanthoMoClo─A Robust Modular Cloning Genetic Toolkit for the Genera Xanthobacter and Roseixanthobacter
Abstract
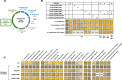
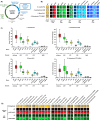
Interest in Xanthobacter species is increasing due to their unique metabolic capabilities. They can grow in both heterotrophic and fully autotrophic environments, including carbon dioxide, dinitrogen gas, and hydrogen as the sole carbon, nitrogen, and energy sources, respectively. Academic and industrial groups looking to leverage these metabolic properties are already using Xanthobacter strains for the sustainable production of food and commodities. However, only a handful of genetic parts and protocols exist in scattered genetic backgrounds, and there is an unmet need for reliable genetic engineering tools to manipulate Xanthobacter species. Here, we developed XanthoMoClo, a robust modular cloning genetic toolkit for Xanthobacter and Roseixanthobacter species and strains, providing extensive tools to transform them, manipulate their metabolism, and express genes of interest. The toolkit contains plasmid parts, such as replication origins, antibiotic selection markers, fluorescent proteins, constitutive and inducible promoters, a standardized framework to incorporate novel components into the toolkit, and a conjugation donor to transform Xanthobacter and Roseixanthobacter strains easily with no or minimal optimization. We validated these plasmid components in depth in three of the most commonly studied Xanthobacter strains: X. versatilis Py2, X. autotrophicus GZ29, and X. flavus GJ10, as well as in R. finlandensis VTT E-85241. Finally, we demonstrate robust toolkit functionality across 21 different species of Xanthobacter and Roseixanthobacter, comprising 23 strains in total. The XanthoMoClo genetic toolkit is available to the research community (through AddGene) and will help accelerate the genetic engineering of Xanthobacter to further their applications in sustainability and bioremediation efforts.
Keywords: MoClo; Xanthobacter; genetic engineering; genetic toolkit; sustainability; synthetic biology.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






References
-
- Wiegel J. K. W.Xanthobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd, 2015, pp. 1–22. DOI: 10.1002/9781118960608.gbm00829. - DOI
-
- Soltysiak M. P. M.; Jalihal A. P.; Christophersen C. E.; Ory A. L. H.; Lee A. D.; Boulton J.; Springer M. Expansion and revision of the genus Xanthobacter and proposal of Roseixanthobacter gen. nov. bioRxiv 2024, 2024.07.11.603134.
-
- Wiegel J. K. W.Xanthobacter Wiegel, Wilke, Baumgarten, Opitz and Schlegel 1978, 573AL. In Bergey’s Manual® of Systematic Bacteriology, Brenner D. J.; Krieg N. R.; Garrity G. M.; Staley J. T.; Boone D. R.; Vos P.; Goodfellow M.; Rainey F. A.; Schleifer K.-H., Eds.; Springer-Verlag: New York, 2005; pp. 555–566. DOI: 10.1007/0-387-29298-5_135. - DOI
-
- Wiegel J.; Wilke D.; Baumgarten J.; Opitz R.; Schlegel H. Transfer of the Nitrogen-Fixing Hydrogen Bacterium Corynebacterium Autotrophicum Baumgarten et al. to Xanthobacter Gen. Nov. Int. J. Syst. Evol. Microbiol. 1978, 28 (4), 573–581. 10.1099/00207713-28-4-573. - DOI
-
- IEA. Renewables 2023, 2024. https://www.iea.org/reports/renewables-2023.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

