Repair pathway coordination from gap filling by polβ and subsequent nick sealing by LIG1 or LIG3α governs BER efficiency at the downstream steps
- PMID: 40081282
- PMCID: PMC12038979
- DOI: 10.1016/j.dnarep.2025.103826
Repair pathway coordination from gap filling by polβ and subsequent nick sealing by LIG1 or LIG3α governs BER efficiency at the downstream steps
Abstract
Base excision repair (BER) is the critical mechanism for preventing mutagenic and lethal consequences of single base lesions generated by endogenous factors or exposure to environmental hazards. BER pathway involves multi-step enzymatic reactions that require a tight coordination between repair proteins to transfer DNA intermediates in an orderly manner. Though often considered an accurate process, the BER can contribute to genome instability if normal coordination between gap filling by DNA polymerase (pol) β and subsequent nick sealing by DNA ligase 1 (LIG1) or DNA ligase 3α (LIG3α) breaks down at the downstream steps. Our studies demonstrated that an inaccurate DNA ligation by LIG1/LIG3α, stemming from an uncoordinated repair with polβ, leads to a range of deviations from canonical BER pathway, faulty repair events, and formation of deleterious DNA intermediates. Furthermore, X-ray repair cross-complementing protein 1 (XRCC1), as a scaffolding factor, enhances the processivity of downstream steps, and the DNA-end processing enzymes, Aprataxin (APTX), Flap-Endonuclease 1 (FEN1), and AP-Endonuclease 1 (APE1), play critical roles for cleaning of ligase failure products and proofreading of polβ errors in coordination with BER ligases. Overall, our studies contribute to understanding of how a multi-protein repair complex interplay at the final steps to maintain the repair efficiency.
Keywords: Base excision repair; Cancer; DNA ligase; DNA polymerase; Genome stability; Repair coordination.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest No competing interests
Figures

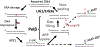
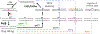

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

