Simultaneous CRISPR screening and spatial transcriptomics reveal intracellular, intercellular, and functional transcriptional circuits
- PMID: 40081369
- PMCID: PMC12135205
- DOI: 10.1016/j.cell.2025.02.012
Simultaneous CRISPR screening and spatial transcriptomics reveal intracellular, intercellular, and functional transcriptional circuits
Abstract
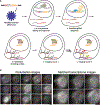
Pooled optical screens have enabled the study of cellular interactions, morphology, or dynamics at massive scale, but they have not yet leveraged the power of highly plexed single-cell resolved transcriptomic readouts to inform molecular pathways. Here, we present a combination of imaging spatial transcriptomics with parallel optical detection of in situ amplified guide RNAs (Perturb-FISH). Perturb-FISH recovers intracellular effects that are consistent with single-cell RNA-sequencing-based readouts of perturbation effects (Perturb-seq) in a screen of lipopolysaccharide response in cultured monocytes, and it uncovers intercellular and density-dependent regulation of the innate immune response. Similarly, in three-dimensional xenograft models, Perturb-FISH identifies tumor-immune interactions altered by genetic knockout. When paired with a functional readout in a separate screen of autism spectrum disorder risk genes in human-induced pluripotent stem cell (hIPSC) astrocytes, Perturb-FISH shows common calcium activity phenotypes and their associated genetic interactions and dysregulated molecular pathways. Perturb-FISH is thus a general method for studying the genetic and molecular associations of spatial and functional biology at single-cell resolution.
Keywords: multimodal screening; pooled CRISPR screen; pooled optical profiling; single-cell perturbations; spatial transcriptomics.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests L.B., B.C., and S.L.F. are inventors on a patent application relating to work described in the manuscript, which has been filed by the Broad Institute.
Figures




Update of
-
Simultaneous CRISPR screening and spatial transcriptomics reveals intracellular, intercellular, and functional transcriptional circuits.bioRxiv [Preprint]. 2023 Dec 1:2023.11.30.569494. doi: 10.1101/2023.11.30.569494. bioRxiv. 2023. Update in: Cell. 2025 Apr 17;188(8):2141-2158.e18. doi: 10.1016/j.cell.2025.02.012. PMID: 38076932 Free PMC article. Updated. Preprint.
References
-
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. (2016). Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17. 10.1016/j.cell.2016.11.038. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

