Bi-allelic MED16 variants cause a MEDopathy with intellectual disability, motor delay, and craniofacial, cardiac, and limb malformations
- PMID: 40081376
- PMCID: PMC12081314
- DOI: 10.1016/j.ajhg.2025.02.016
Bi-allelic MED16 variants cause a MEDopathy with intellectual disability, motor delay, and craniofacial, cardiac, and limb malformations
Abstract
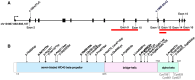
The Mediator complex regulates protein-coding gene transcription by coordinating the interaction of upstream enhancers with the basal transcription machinery at the promoter. Pathogenic variants in Mediator subunits typically lead to neurodevelopmental or neurodegenerative disorders with variable clinical presentations, designated as MEDopathies. Here, we report the identification of 25 individuals from 18 families with bi-allelic MED16 variants who have a multiple congenital anomalies (MCAs)-intellectual disability syndrome. Intellectual disability, speech delay, and/or motor delay of variable severity were constant and associated with variable combinations of craniofacial defects (micro/retrognathia, cleft palate, and preauricular tags), anomalies of the extremities, and heart defects (predominantly tetralogy of Fallot). Visual impairment, deafness, and magnetic resonance imaging (MRI) abnormalities were also frequent. The 26 variants identified were comprised of eight predicted protein-truncating (three intragenic deletions, two frameshifts, and one nonsense and two essential splice site variants) and 18 missense or in-frame duplication variants affecting conserved residues, without clear correlation between phenotypic severity and variant type combination. Three-dimensional modeling indicated that the missense and duplication variants likely have a destabilizing effect on the structural elements of the protein. Immunofluorescence assays demonstrated protein mislocalization from the nucleus to the cytoplasm for 16 of the 17 variants studied. Homozygous mutant med16 zebrafish presented growth delay and increased mortality compared with wild-type fish, and Med16 knockout mice are preweaning lethal, highlighting the conserved requirement of MED16 for development. Overall, we describe an autosomal recessive MCAs-intellectual disability MEDopathy, emphasizing the importance of Mediator during neurodevelopment and suggesting that some tissues are particularly sensitive to the loss of certain subunits.
Keywords: MED16; MEDopathies; Mediator complex; multiple congenital anomalies-intellectual disability syndrome.
Copyright © 2025 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

