Placenta-derived factors contribute to human iPSC-liver organoid growth
- PMID: 40082402
- PMCID: PMC11906828
- DOI: 10.1038/s41467-025-57551-w
Placenta-derived factors contribute to human iPSC-liver organoid growth
Abstract
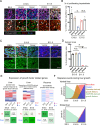
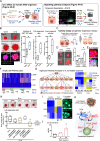
Organoids derived from human induced pluripotent stem cells (hiPSC) are potentially applicable for regenerative medicine. However, the applications have been hampered by limited organoid size and function as a consequence of a lack of progenitor expansion. Here, we report the recapitulation of progenitor expansion in hiPSC-liver organoids based on the analysis of mouse development. Visualization of blood perfusion and oxygen levels in mouse embryos reveals a transient hypoxic environment during hepatoblast expansion, despite active blood flow. During this specific stage, the placenta expresses various growth factors. Human and mouse placenta-liver interaction analysis identifies various placenta-derived factors. Among them, IL1α efficiently induces the growth in hiPSC-liver organoids as well as mouse fetal livers following progenitor expansion under hypoxia. Furthermore, subsequent oxygenation demonstrates that progenitors expanded by IL1α contribute to hiPSC-liver organoid size and function. Taken together, we demonstrate that treatment with the placenta-derived factor under hypoxia is a crucial human organoid culture technique that efficiently induces progenitor expansion.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Takebe, T. et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell16, 556–565 (2015). - PubMed
-
- Takebe, T. et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature499, 481–484 (2013). - PubMed
-
- Camp, J. G. et al. Multilineage communication regulates human liver bud development from pluripotency. Nature546, 533–538 (2017). - PubMed
-
- Amabile, G. & Meissner, A. Induced pluripotent stem cells: current progress and potential for regenerative medicine. Trends Mol. Med.15, 59–68 (2009). - PubMed
-
- Yamanaka, S. Pluripotent stem cell-based cell therapy—promise and challenges. Cell Stem Cell27, 523–531 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- JP23bm1223007/Japan Agency for Medical Research and Development (AMED)
- JP22bm0304002h0310/Japan Agency for Medical Research and Development (AMED)
- JP13bm0304002/Japan Agency for Medical Research and Development (AMED)
- 21H04830/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- 22K16440/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
LinkOut - more resources
Full Text Sources

