NSUN2-mediated m5C modification drives alternative splicing reprogramming and promotes multidrug resistance in anaplastic thyroid cancer through the NSUN2/SRSF6/UAP1 signaling axis
- PMID: 40083919
- PMCID: PMC11898302
- DOI: 10.7150/thno.104713
NSUN2-mediated m5C modification drives alternative splicing reprogramming and promotes multidrug resistance in anaplastic thyroid cancer through the NSUN2/SRSF6/UAP1 signaling axis
Abstract
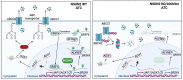
Rationale: Anaplastic thyroid carcinoma (ATC) is an extraordinarily aggressive form of thyroid cancer, frequently presenting with locally advanced infiltration or distant metastases at the time of initial diagnosis, thus missing the optimal window for surgical intervention. Consequently, systemic chemotherapy and targeted therapies are vital for improving the prognosis of ATC. However, ATC exhibits significant resistance to conventional treatments, highlighting the need to elucidate the biological mechanisms underlying this drug resistance and identify novel therapeutic targets to overcome it. Methods: We conducted a comprehensive analysis of both bulk and single-cell RNA sequencing (scRNA-seq) data from ATC samples to screen for m5C modification-related genes associated with multidrug resistance (MDR). We then performed IC50 assays, flow cytometry, and employed a spontaneous tumorigenic ATC mouse model with Nsun2 knockout to demonstrate that NSUN2 promotes MDR in ATC. To investigate the mechanisms of NSUN2-mediated drug resistance, we generated NSUN2-knockout ATC cell lines and performed transcriptomic, proteomic, and MeRIP-seq analyses. Additionally, RNA sequencing and alternative splicing analyses were conducted to determine global changes upon NSUN2 knockout. We further explored the underlying mechanisms of the NSUN2/SRSF6/UAP1 axis through glycoprotein staining, denaturing IP ubiquitination, nuclear-cytoplasmic fractionation, and PCR. Lastly, we evaluated the synergistic effects of a small-molecule NSUN2 inhibitor with anticancer agents both in vitro and in vivo. Results: Our findings reveal that NSUN2 expression correlates significantly with MDR in ATC. NSUN2 operates as a "writer" and ALYREF as a "reader" of m5C on SRSF6 mRNA, inducing alternative splicing reprogramming and redirecting the splice form of the UAP1 gene from AGX1 to AGX2. As a result, AGX2 enhances the N-linked glycosylation of ABC transporters, stabilizing them by preventing ubiquitination-mediated degradation. Furthermore, an NSUN2 inhibitor reduces NSUN2 enzymatic activity and diminishes downstream target expression, presenting a novel, promising therapeutic approach to overcome MDR in ATC. Conclusions: These findings suggest that the NSUN2/SRSF6/UAP1 signaling axis plays a vital role in MDR of ATC and identify NSUN2 as a synergistic target for chemotherapy and targeted therapy in ATC.
Keywords: 5-methylcytosine; NSUN2; alternative splicing reprogramming; anaplastic thyroid cancer; multidrug resistance.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures








References
-
- Abbasifarid E, Sajjadi-Jazi SM, Beheshtian M, Samimi H, Larijani B, Haghpanah V. The Role of ATP-Binding Cassette Transporters in the Chemoresistance of Anaplastic Thyroid Cancer: A Systematic Review. Endocrinology. 2019;160:2015–23. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

